Dement Neurocogn Disord.
2016 Sep;15(3):61-67. 10.12779/dnd.2016.15.3.61.
Apolipoprotein E4: A Risk Factor for Successful Cognitive Aging
- Affiliations
-
- 1Department of Neurology, School of Medicine, Catholic University of Daegu, Daegu, Korea. dolbaeke@cu.ac.kr
- KMID: 2442847
- DOI: http://doi.org/10.12779/dnd.2016.15.3.61
Abstract
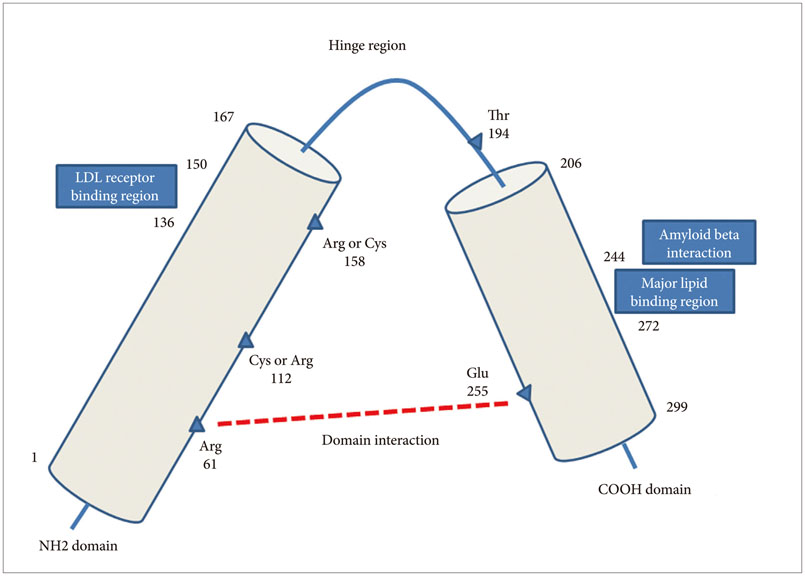
- Apolipoprotein E is a plasma protein that has an important role in transport and metabolism of lipids in serum as well as central nervous system. Among the 3 common alleles, the ε2 allele has the most stable structure followed by ε3 and ε4 in order. There is evidence for a deleterious role of ε4 allele by atherosclerosis and amyloid beta accumulation in brain and body. The presence and gene dose of ε4 allele are risk factors for late-onset Alzheimer's disease. Apolipoprotein E ε4 may have a role in the pathology of amyloid beta and tau and it has a strong relationship with the early onset of late-onset Alzheimer's disease. However, early-onset Alzheimer's disease has a weaker relationship with ε4 allele of apolipoprotein E.
Keyword
MeSH Terms
Figure
Reference
-
1. Corbo RM, Prévost M, Raussens V, Gambina G, Moretto G, Scacchi R. Structural and phylogenetic approaches to assess the significance of human Apolipoprotein E variation. Mol Genet Metab. 2006; 89:261–269.
Article2. Nickerson DA, Taylor SL, Fullerton SM, Weiss KM, Clark AG, Stengård JH, et al. Sequence diversity and large-scale typing of SNPs in the human apolipoprotein E gene. Genome Res. 2000; 10:1532–1545.
Article3. Park CH, Lee ST, Ki CS, Kim JW. Discrepancy in genotyping of apolipoprotein E between allele-specific PCR and fluorescence resonance energy transfer or sequencing. Korean J Lab Med. 2010; 30:325–328.
Article4. Scacchi R, Gambina G, Ferrari G, Corbo RM. Screening of two mutations at exon 3 of the apolipoprotein E gene (sites 28 and 42) in a sample of patients with sporadic late-onset Alzheimer's disease. Neurobiol Aging. 2003; 24:339–343.
Article5. Wernette-Hammond ME, Lauer SJ, Corsini A, Walker D, Taylor JM, Rall SC Jr. Glycosylation of human apolipoprotein E. The carbohydrate attachment site is threonine 194. J Biol Chem. 1989; 264:9094–9101.
Article6. Weisgraber KH. Apolipoprotein E: structure-function relationships. Adv Protein Chem. 1994; 45:249–302.
Article7. Mahley RW, Huang Y. Apolipoprotein e sets the stage: response to injury triggers neuropathology. Neuron. 2012; 76:871–885.
Article8. Mahley RW, Huang Y. Apolipoprotein E: from atherosclerosis to Alzheimer's disease and beyond. Curr Opin Lipidol. 1999; 10:207–217.
Article9. Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer's disease to AIDS. J Lipid Res. 2009; 50:Suppl. S183–S188.
Article10. Zhang H, Wu LM, Wu J. Cross-talk between apolipoprotein E and cytokines. Mediators Inflamm. 2011; 2011:949072.
Article11. Kim J, Yoon H, Basak J, Kim J. Apolipoprotein E in synaptic plasticity and Alzheimer's disease: potential cellular and molecular mechanisms. Mol Cells. 2014; 37:767–776.
Article12. Xu Q, Bernardo A, Walker D, Kanegawa T, Mahley RW, Huang Y. Profile and regulation of apolipoprotein E (ApoE) expression in the CNS in mice with targeting of green fluorescent protein gene to the ApoE locus. J Neurosci. 2006; 26:4985–4994.
Article13. Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993; 261:921–923.
Article14. Rebeck GW, Kindy M, LaDu MJ. Apolipoprotein E and Alzheimer's disease: the protective effects of ApoE2 and E3. J Alzheimers Dis. 2002; 4:145–154.
Article15. Villeneuve S, Brisson D, Marchant NL, Gaudet D. The potential applications of Apolipoprotein E in personalized medicine. Front Aging Neurosci. 2014; 6:154.
Article16. Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997; 278:1349–1356.
Article17. van Duijn CM, de Knijff P, Cruts M, Wehnert A, Havekes LM, Hofman A, et al. Apolipoprotein E4 allele in a population-based study of early-onset Alzheimer's disease. Nat Genet. 1994; 7:74–78.
Article18. Kwon OD, Khaleeq A, Chan W, Pavlik VN, Doody RS. Apolipoprotein E polymorphism and age at onset of Alzheimer's disease in a quadriethnic sample. Dement Geriatr Cogn Disord. 2010; 30:486–491.
Article19. Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC Jr, et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet. 1994; 7:180–184.
Article20. Wu YN, Zhang JW, Zhang ZX, Qu QM, Chen D. [An association analysis of apolipoprotein E genotypes with Alzheimer's disease in Chinese population]. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2001; 23:450–454.
Article21. Mahley RW, Huang Y. Alzheimer disease: multiple causes, multiple effects of apolipoprotein E4, and multiple therapeutic approaches. Ann Neurol. 2009; 65:623–625.
Article22. Raffai RL, Dong LM, Farese RV Jr, Weisgraber KH. Introduction of human apolipoprotein E4 "domain interaction" into mouse apolipoprotein E. Proc Natl Acad Sci U S A. 2001; 98:11587–11591.
Article23. Ye S, Huang Y, Müllendorff K, Dong L, Giedt G, Meng EC, et al. Apolipoprotein (apo) E4 enhances amyloid beta peptide production in cultured neuronal cells: apoE structure as a potential therapeutic target. Proc Natl Acad Sci U S A. 2005; 102:18700–18705.
Article24. Hatters DM, Peters-Libeu CA, Weisgraber KH. Apolipoprotein E structure: insights into function. Trends Biochem Sci. 2006; 31:445–454.
Article25. Rohn TT. Proteolytic cleavage of apolipoprotein E4 as the keystone for the heightened risk associated with Alzheimer's disease. Int J Mol Sci. 2013; 14:14908–14922.
Article26. Rohn TT, Catlin LW, Coonse KG, Habig JW. Identification of an amino-terminal fragment of apolipoprotein E4 that localizes to neurofibrillary tangles of the Alzheimer's disease brain. Brain Res. 2012; 1475:106–115.
Article27. Chang S, ran Ma T, Miranda RD, Balestra ME, Mahley RW, Huang Y. Lipid- and receptor-binding regions of apolipoprotein E4 fragments act in concert to cause mitochondrial dysfunction and neurotoxicity. Proc Natl Acad Sci U S A. 2005; 102:18694–18699.
Article28. Rubinsztein DC. The genetics of Alzheimer's disease. Prog Neurobiol. 1997; 52:447–454.
Article29. Tanzi RE. The genetics of Alzheimer disease. Cold Spring Harb Perspect Med. 2012; 2(10):pii: a006296. DOI: 10.1101/cshperspect.a006296.
Article30. Raichlen DA, Alexander GE. Exercise, APOE genotype, and the evolution of the human lifespan. Trends Neurosci. 2014; 37:247–255.
Article31. Golde TE, Eckman CB, Younkin SG. Biochemical detection of Abeta isoforms: implications for pathogenesis, diagnosis, and treatment of Alzheimer's disease. Biochim Biophys Acta. 2000; 1502:172–187.
Article32. Lee VM, Trojanowski JQ. The disordered neuronal cytoskeleton in Alzheimer's disease. Curr Opin Neurobiol. 1992; 2:653–656.
Article33. Golde TE, Petrucelli L, Lewis J. Targeting Abeta and tau in Alzheimer's disease, an early interim report. Exp Neurol. 2010; 223:252–266.34. Selkoe DJ. Clearing the brain's amyloid cobwebs. Neuron. 2001; 32:177–180.35. Selkoe DJ. Alzheimer's disease: genes, proteins, and therapy. Physiol Rev. 2001; 81:741–766.
Article36. Vemuri P, Wiste HJ, Weigand SD, Knopman DS, Shaw LM, Trojanowski JQ, et al. Effect of apolipoprotein E on biomarkers of amyloid load and neuronal pathology in Alzheimer disease. Ann Neurol. 2010; 67:308–316.
Article37. Harris FM, Brecht WJ, Xu Q, Mahley RW, Huang Y. Increased tau phosphorylation in apolipoprotein E4 transgenic mice is associated with activation of extracellular signal-regulated kinase: modulation by zinc. J Biol Chem. 2004; 279:44795–44801.
Article38. Tesseur I, Van Dorpe J, Spittaels K, Van den Haute C, Moechars D, Van Leuven F. Expression of human apolipoprotein E4 in neurons causes hyperphosphorylation of protein tau in the brains of transgenic mice. Am J Pathol. 2000; 156:951–964.
Article39. Agosta F, Vossel KA, Miller BL, Migliaccio R, Bonasera SJ, Filippi M, et al. Apolipoprotein E epsilon4 is associated with disease-specific effects on brain atrophy in Alzheimer's disease and frontotemporal dementia. Proc Natl Acad Sci U S A. 2009; 106:2018–2022.
Article40. Stancu IC, Vasconcelos B, Terwel D, Dewachter I. Models of β-amyloid induced Tau-pathology: the long and "folded" road to understand the mechanism. Mol Neurodegener. 2014; 9:51.
Article41. Inbar D, Belinson H, Rosenman H, Michaelson DM. Possible role of tau in mediating pathological effects of apoE4 in vivo prior to and following activation of the amyloid cascade. Neurodegener Dis. 2010; 7:16–23.
Article42. Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002; 297:353–356.
Article43. Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991; 82:239–259.
Article44. Frisoni GB, Ganzola R, Canu E, Rüb U, Pizzini FB, Alessandrini F, et al. Mapping local hippocampal changes in Alzheimer's disease and normal ageing with MRI at 3 Tesla. Brain. 2008; 131(Pt 12):3266–3276.
Article45. Sabri O, Sabbagh MN, Seibyl J, Barthel H, Akatsu H, Ouchi Y, et al. Florbetaben PET imaging to detect amyloid beta plaques in Alzheimer's disease: phase 3 study. Alzheimers Dement. 2015; 11:964–974.
Article46. Rusinek H, De Santi S, Frid D, Tsui WH, Tarshish CY, Convit A, et al. Regional brain atrophy rate predicts future cognitive decline: 6-year longitudinal MR imaging study of normal aging. Radiology. 2003; 229:691–696.
Article47. McGowan E, Pickford F, Kim J, Onstead L, Eriksen J, Yu C, et al. Abeta42 is essential for parenchymal and vascular amyloid deposition in mice. Neuron. 2005; 47:191–199.48. Mori E, Lee K, Yasuda M, Hashimoto M, Kazui H, Hirono N, et al. Accelerated hippocampal atrophy in Alzheimer's disease with apolipoprotein E epsilon4 allele. Ann Neurol. 2002; 51:209–214.49. Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993; 90:1977–1981.
Article50. Strittmatter WJ, Saunders AM, Goedert M, Weisgraber KH, Dong LM, Jakes R, et al. Isoform-specific interactions of apolipoprotein E with microtubule-associated protein tau: implications for Alzheimer disease. Proc Natl Acad Sci U S A. 1994; 91:11183–11186.
Article51. Sunderland T, Mirza N, Putnam KT, Linker G, Bhupali D, Durham R, et al. Cerebrospinal fluid beta-amyloid1-42 and tau in control subjects at risk for Alzheimer's disease: the effect of APOE epsilon4 allele. Biol Psychiatry. 2004; 56:670–676.
Article52. Walsh DM, Selkoe DJ. Deciphering the molecular basis of memory failure in Alzheimer's disease. Neuron. 2004; 44:181–193.
Article53. Conforti L, Adalbert R, Coleman MP. Neuronal death: where does the end begin? Trends Neurosci. 2007; 30:159–166.
Article54. Rhinn H, Fujita R, Qiang L, Cheng R, Lee JH, Abeliovich A. Integrative genomics identifies APOE ε4 effectors in Alzheimer's disease. Nature. 2013; 500:45–50.
Article55. Desikan RS, Thompson WK, Holland D, Hess CP, Brewer JB, Zetterberg H, et al. The role of clusterin in amyloid-β-associated neurodegeneration. JAMA Neurol. 2014; 71:180–187.
Article56. Albuquerque MS, Mahar I, Davoli MA, Chabot JG, Mechawar N, Quirion R, et al. Regional and sub-regional differences in hippocampal GABAergic neuronal vulnerability in the TgCRND8 mouse model of Alzheimer's disease. Front Aging Neurosci. 2015; 7:30.
Article57. Goutagny R, Krantic S. Hippocampal oscillatory activity in Alzheimer's disease: toward the identification of early biomarkers? Aging Dis. 2013; 4:134–140.
Article58. Roberson ED, Scearce-Levie K, Palop JJ, Yan F, Cheng IH, Wu T, et al. Reducing endogenous tau ameliorates amyloid beta-induced deficits in an Alzheimer's disease mouse model. Science. 2007; 316:750–754.
Article59. Seward ME, Swanson E, Norambuena A, Reimann A, Cochran JN, Li R, et al. Amyloid-β signals through tau to drive ectopic neuronal cell cycle re-entry in Alzheimer's disease. J Cell Sci. 2013; 126(Pt 5):1278–1286.
Article60. Lane-Donovan C, Philips GT, Herz J. More than cholesterol transporters: lipoprotein receptors in CNS function and neurodegeneration. Neuron. 2014; 83:771–787.
Article61. Krstic D, Knuesel I. Deciphering the mechanism underlying late-onset Alzheimer disease. Nat Rev Neurol. 2013; 9:25–34.
Article62. Kim DH, Yeo SH, Park JM, Choi JY, Lee TH, Park SY, et al. Genetic markers for diagnosis and pathogenesis of Alzheimer's disease. Gene. 2014; 545:185–193.
Article63. Leduc V, Domenger D, De Beaumont L, Lalonde D, Bélanger-Jasmin S, Poirier J. Function and comorbidities of apolipoprotein e in Alzheimer's disease. Int J Alzheimers Dis. 2011; 2011:974361.64. D'Onofrio G, Sancarlo D, Panza F, Copetti M, Cascavilla L, Paris F, et al. Neuropsychiatric symptoms and functional status in Alzheimer's disease and vascular dementia patients. Curr Alzheimer Res. 2012; 9:759–771.65. Chen CH, Mizuno T, Elston R, Kariuki MM, Hall K, Unverzagt F, et al. A comparative study to screen dementia and APOE genotypes in an ageing East African population. Neurobiol Aging. 2010; 31:732–740.
Article66. Mawuenyega KG, Sigurdson W, Ovod V, Munsell L, Kasten T, Morris JC, et al. Decreased clearance of CNS beta-amyloid in Alzheimer's disease. Science. 2010; 330:1774.
Article67. Castellano JM, Kim J, Stewart FR, Jiang H, DeMattos RB, Patterson BW, et al. Human apoE isoforms differentially regulate brain amyloid-β peptide clearance. Sci Transl Med. 2011; 3:89ra57.
Article68. Tang MX, Stern Y, Marder K, Bell K, Gurland B, Lantigua R, et al. The APOE-epsilon4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. JAMA. 1998; 279:751–755.
Article69. Hendrie HC, Murrell J, Gao S, Unverzagt FW, Ogunniyi A, Hall KS. International studies in dementia with particular emphasis on populations of African origin. Alzheimer Dis Assoc Disord. 2006; 20:3 Suppl2. S42–S46.
Article70. Evans DA, Bennett DA, Wilson RS, Bienias JL, Morris MC, Scherr PA, et al. Incidence of Alzheimer disease in a biracial urban community: relation to apolipoprotein E allele status. Arch Neurol. 2003; 60:185–189.
Article71. Holtzman DM. CSF biomarkers for Alzheimer's disease: current utility and potential future use. Neurobiol Aging. 2011; 32:Suppl 1. S4–S9.
Article72. Alzheimer A. Die Seelenstorungen auf arteriscleroticsher Grundlage. Allgem Z Psychiatr Psych Gerichtl Med. 1902; 59:695–711.
Article73. Weggen S, Beher D. Molecular consequences of amyloid precursor protein and presenilin mutations causing autosomal-dominant Alzheimer's disease. Alzheimers Res Ther. 2012; 4:9.
Article74. Cataldo AM, Barnett JL, Pieroni C, Nixon RA. Increased neuronal endocytosis and protease delivery to early endosomes in sporadic Alzheimer's disease: neuropathologic evidence for a mechanism of increased beta-amyloidogenesis. J Neurosci. 1997; 17:6142–6151.
Article75. Cataldo AM, Peterhoff CM, Troncoso JC, Gomez-Isla T, Hyman BT, Nixon RA. Endocytic pathway abnormalities precede amyloid beta deposition in sporadic Alzheimer's disease and Down syndrome: differential effects of APOE genotype and presenilin mutations. Am J Pathol. 2000; 157:277–286.
Article76. Fullerton SM, Clark AG, Weiss KM, Nickerson DA, Taylor SL, Stengârd JH, et al. Apolipoprotein E variation at the sequence haplotype level: implications for the origin and maintenance of a major human polymorphism. Am J Hum Genet. 2000; 67:881–900.
Article77. McIntosh AM, Bennett C, Dickson D, Anestis SF, Watts DP, Webster TH, et al. The apolipoprotein E (APOE) gene appears functionally monomorphic in chimpanzees (Pan troglodytes). PLoS One. 2012; 7:e47760.
Article78. Finch CE, Stanford CB. Meat-adaptive genes and the evolution of slower aging in humans. Q Rev Biol. 2004; 79:3–50.
Article79. Eisenberg DT, Kuzawa CW, Hayes MG. Worldwide allele frequencies of the human apolipoprotein E gene: climate, local adaptations, and evolutionary history. Am J Phys Anthropol. 2010; 143:100–111.80. Kwon OD, Choi SY, Park JH, Yoon CH, Kwon HH, Shin IH. Association of apolipoprotein E gene polymorphism with cognitive function of the elderly residents in a rural community. J Korean Neurol Assoc. 2009; 27:362–368.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Interaction of Apolipoprotein E epsilon4 and Education on Cognitive Decline in Korean Elders
- Genetic and Neuropsychological Factor Analysis on the Cognitive Function of the Elderly Population: APOE Genotype and Age Effect Analysis
- Apolipoprotein E4 Genotype and Depressive Symptoms as Risk Factors for Dementia in an Older Korean Population
- Apolipoprotein E Polymorphism and Cholesterol in Stones and Bile of Patients with Cholelithiasis
- Apolipoprotein E Polymorphism and Cognitive Function Change of the Elderly in a Rural Area, Korea