J Breast Cancer.
2017 Sep;20(3):246-253. 10.4048/jbc.2017.20.3.246.
Copy Number Profiling of MammaPrintâ„¢ Genes Reveals Association with the Prognosis of Breast Cancer Patients
- Affiliations
-
- 1Cancer Genetics and Epigenetics Lab, Department of Biosciences, COMSATS Institute of Information Technology, Islamabad, Pakistan. farhan.haq@comsats.edu.pk
- 2Department of Pharmacology & Immune Network Pioneer Research Center, Ajou University School of Medicine, Suwon, Korea.
- KMID: 2438991
- DOI: http://doi.org/10.4048/jbc.2017.20.3.246
Abstract
- PURPOSE
The MammaPrintâ„¢ gene signature, currently used in clinical practice, provides prognostic information regarding the recurrence and potential metastasis in breast cancer patients. However, the prognostic information of the 70 genes included can only be estimated at the RNA expression level. In this study, we investigated whether copy number information of MammaPrintâ„¢ genes at the DNA level can be used as a prognostic tool for breast cancer, as copy number variations (CNVs) are major contributors to cancer progression.
METHODS
We performed CNV profiling of MammaPrintâ„¢ genes in 59 breast cancer cell lines and 650 breast cancer patients, using publicly available data in The Cancer Genome Atlas (TCGA) database. Statistical analyses including Fisher exact test, chi-square test, and Kaplan-Meier survival analyses were performed.
RESULTS
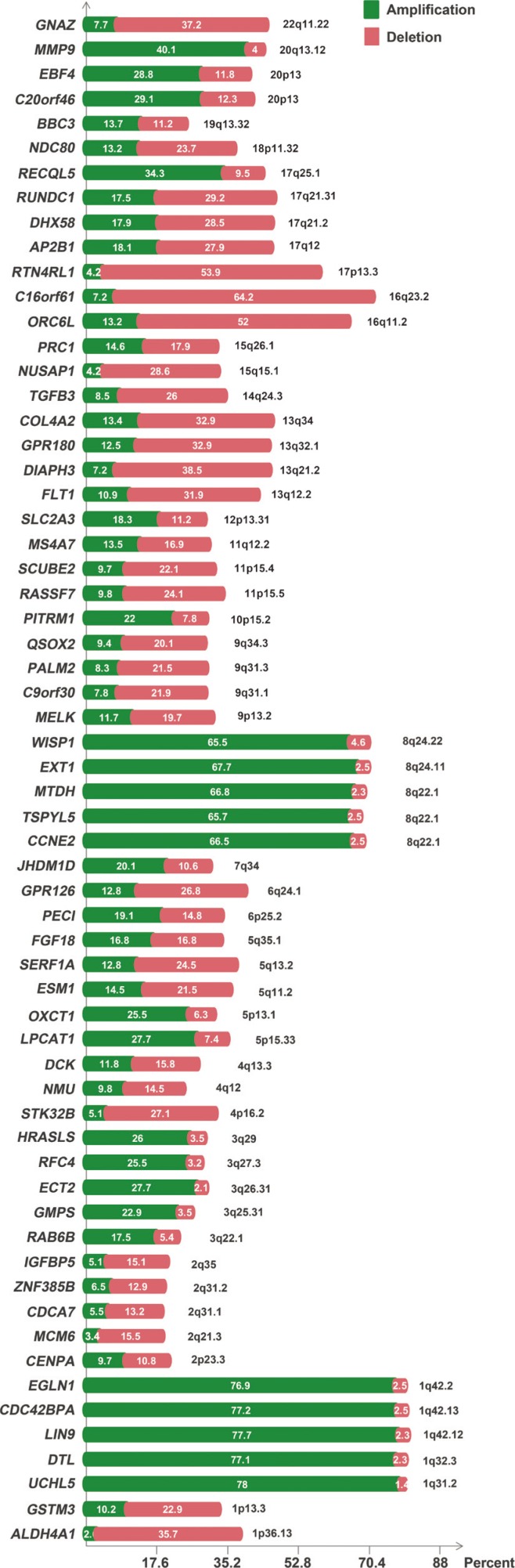
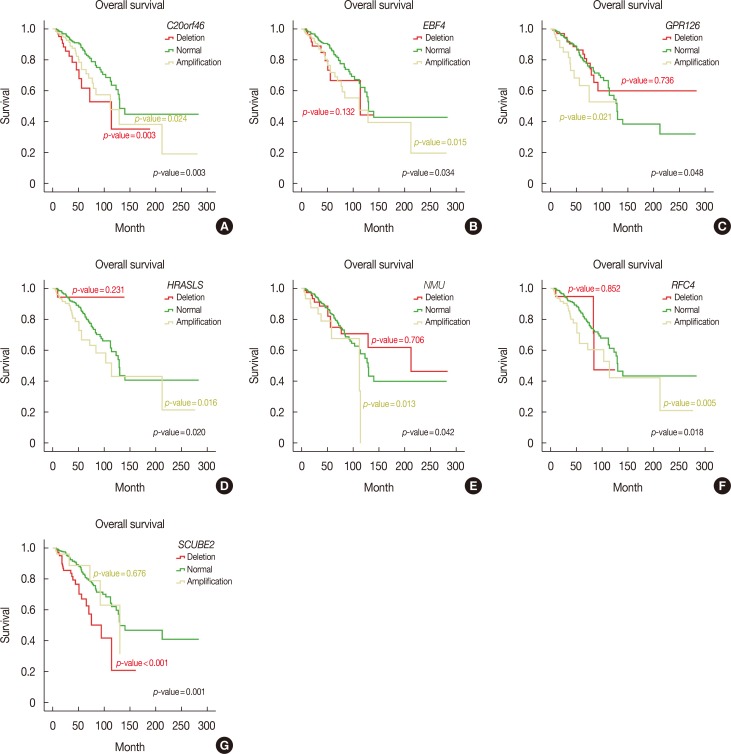
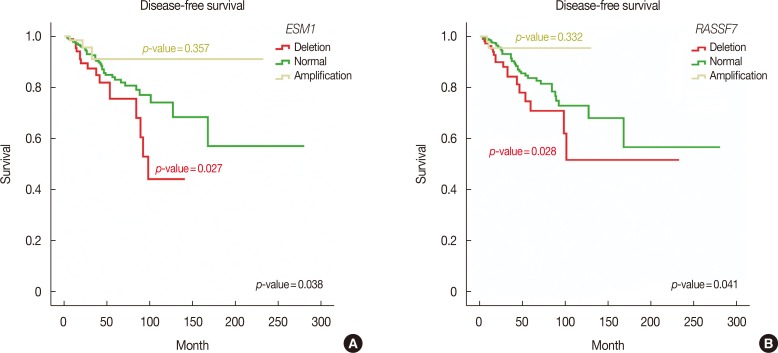
All MammaPrintâ„¢ genes showed recurrent CNVs, particularly in TCGA cohort. CNVs of 32 and 36 genes showed significant associations with progesterone receptor and estrogen rector, respectively. No genes showed a significant association with human epidermal growth factor receptor 2 status and lymph node status. In addition, only six genes were associated with tumor stages. RFC4, HRASLS, NMU, GPR126, SCUBE2, C20orf46, and EBF4 were associated with reduced survival and RASSF7 and ESM1 were associated with reduced disease-free survival.
CONCLUSION
Based on these findings, a concordance of CNV-based genomic rearrangement with expression profiling of these genes and their putative roles in disease tumorigenesis was established. The results suggested that the CNV profiles of the MammaPrintâ„¢ genes can be used to predict the prognosis of breast cancer patients. In addition, this approach may lead to the development of new cancer biomarkers at the DNA level.
MeSH Terms
-
Biomarkers
Biomarkers, Tumor
Breast Neoplasms*
Breast*
Carcinogenesis
Cell Line
Cohort Studies
Disease-Free Survival
DNA
DNA Copy Number Variations
Estrogens
Genome
Humans
Lymph Nodes
Neoplasm Metastasis
Prognosis*
Receptor, Epidermal Growth Factor
Receptors, Progesterone
Recurrence
RNA
Biomarkers
Biomarkers, Tumor
DNA
Estrogens
RNA
Receptor, Epidermal Growth Factor
Receptors, Progesterone
Figure
Reference
-
1. Tian S, Roepman P, Van't Veer LJ, Bernards R, de Snoo F, Glas AM. Biological functions of the genes in the mammaprint breast cancer profile reflect the hallmarks of cancer. Biomark Insights. 2010; 5:129–138. PMID: 21151591.
Article2. Ellsworth RE, Decewicz DJ, Shriver CD, Ellsworth DL. Breast cancer in the personal genomics era. Curr Genomics. 2010; 11:146–161. PMID: 21037853.3. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012; 490:61–70. PMID: 23000897.4. Sharma GN, Dave R, Sanadya J, Sharma P, Sharma KK. Various types and management of breast cancer: an overview. J Adv Pharm Technol Res. 2010; 1:109–126. PMID: 22247839.5. Hwang KT, Han W, Cho J, Lee JW, Ko E, Kim EK, et al. Genomic copy number alterations as predictive markers of systemic recurrence in breast cancer. Int J Cancer. 2008; 123:1807–1815. PMID: 18649361.
Article6. Dai X, Xiang L, Li T, Bai Z. Cancer hallmarks, biomarkers and breast cancer molecular subtypes. J Cancer. 2016; 7:1281–1294. PMID: 27390604.
Article7. Onitilo AA, Engel JM, Greenlee RT, Mukesh BN. Breast cancer subtypes based on ER/PR and Her2 expression: comparison of clinicopathologic features and survival. Clin Med Res. 2009; 7:4–13. PMID: 19574486.
Article8. Verigos J, Magklara A. Revealing the complexity of breast cancer by next generation sequencing. Cancers (Basel). 2015; 7:2183–2200. PMID: 26561834.
Article9. Deming SL, Ren Z, Cai Q, Shu XO, Wen W, Long JR, et al. IGF-I and IGF-II genetic variation and breast cancer risk in Chinese women: results from the Shanghai Breast Cancer Study. Cancer Epidemiol Biomarkers Prev. 2008; 17:255–257. PMID: 18199734.
Article10. Hwangbo W, Lee JH, Ahn S, Kim S, Park KH, Kim CH, et al. EGFR gene amplification and protein expression in invasive ductal carcinoma of the breast. Korean J Pathol. 2013; 47:107–115. PMID: 23667369.11. Chen Y, Hao J, Jiang W, He T, Zhang X, Jiang T, et al. Identifying potential cancer driver genes by genomic data integration. Sci Rep. 2013; 3:3538. PMID: 24346768.
Article12. Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012; 486:400–404. PMID: 22722201.
Article13. Urquidi V, Goodison S. Genomic signatures of breast cancer metastasis. Cytogenet Genome Res. 2007; 118:116–129. PMID: 18000362.
Article14. Győrffy B, Hatzis C, Sanft T, Hofstatter E, Aktas B, Pusztai L. Multigene prognostic tests in breast cancer: past, present, future. Breast Cancer Res. 2015; 17:11. PMID: 25848861.
Article15. Ping Z, Siegal GP, Almeida JS, Schnitt SJ, Shen D. Mining genome sequencing data to identify the genomic features linked to breast cancer histopathology. J Pathol Inform. 2014; 5:3. PMID: 24672738.
Article16. Wittner BS, Sgroi DC, Ryan PD, Bruinsma TJ, Glas AM, Male A, et al. Analysis of the MammaPrint breast cancer assay in a predominantly postmenopausal cohort. Clin Cancer Res. 2008; 14:2988–2993. PMID: 18483364.
Article17. Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012; 483:603–607. PMID: 22460905.18. Esposito A, Criscitiello C, Curigliano G. Highlights from the 14(th) St Gallen International Breast Cancer Conference 2015 in Vienna: dealing with classification, prognostication, and prediction refinement to personalize the treatment of patients with early breast cancer. Ecancermedicalscience. 2015; 9:518. PMID: 25932042.
Article19. Shlien A, Malkin D. Copy number variations and cancer. Genome Med. 2009; 1:62. PMID: 19566914.
Article20. Ziogas D, Roukos DH. Genetics and personal genomics for personalized breast cancer surgery: progress and challenges in research and clinical practice. Ann Surg Oncol. 2009; 16:1771–1782. PMID: 19322611.
Article21. Jang H, Hur Y, Lee H. Identification of cancer-driver genes in focal genomic alterations from whole genome sequencing data. Sci Rep. 2016; 6:25582. PMID: 27156852.
Article22. Eirew P, Steif A, Khattra J, Ha G, Yap D, Farahani H, et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature. 2015; 518:422–426. PMID: 25470049.
Article23. Li J, Wang J, Chen Y, Yang L, Chen S. A prognostic 4-gene expression signature for squamous cell lung carcinoma. J Cell Physiol. 2017; 2. 04. Epub. DOI: 10.1002/jcp.25846.
Article24. Liberg D, Sigvardsson M, Akerblad P. The EBF/Olf/Collier family of transcription factors: regulators of differentiation in cells originating from all three embryonal germ layers. Mol Cell Biol. 2002; 22:8389–8397. PMID: 12446759.
Article25. Xiang J, Fang L, Luo Y, Yang Z, Liao Y, Cui J, et al. Levels of human replication factor C4, a clamp loader, correlate with tumor progression and predict the prognosis for colorectal cancer. J Transl Med. 2014; 12:320. PMID: 25407051.
Article26. Cui H, Wang Y, Huang H, Yu W, Bai M, Zhang L, et al. GPR126 protein regulates developmental and pathological angiogenesis through modulation of VEGFR2 receptor signaling. J Biol Chem. 2014; 289:34871–34885. PMID: 25217645.
Article27. Maiga A, Lemieux S, Pabst C, Lavallée VP, Bouvier M, Sauvageau G, et al. Transcriptome analysis of G protein-coupled receptors in distinct genetic subgroups of acute myeloid leukemia: identification of potential disease-specific targets. Blood Cancer J. 2016; 6:e431. PMID: 27258612.
Article28. Song Q, Li C, Feng X, Yu A, Tang H, Peng Z, et al. Decreased expression of SCUBE2 is associated with progression and prognosis in colorectal cancer. Oncol Rep. 2015; 33:1956–1964. PMID: 25672935.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Association between Mitochondrial D-loop Polymorphism and Copy Number
- Mitochondrial Ribosomal Protein S17 Silencing Inhibits Proliferation and Invasiveness of Lung Cancer Cells
- The genomic landscape associated with resistance to aromatase inhibitors in breast cancer
- Clinical Significance of MET Gene Copy Number in Patients with Curatively Resected Gastric Cancer
- Classification of Genes Based on Age-Related Differential Expression in Breast Cancer