Cancer Res Treat.
2019 Jan;51(1):128-140. 10.4143/crt.2017.598.
Prevalence and Prognostic Role of PIK3CA/AKT1 Mutations in Chinese Breast Cancer Patients
- Affiliations
-
- 1Laboratory of Molecular Diagnosis of Cancer, Clinical Research Center for Breast, State Key Laboratory of Biotherapy, National Collaborative Innovation Center for Biotherapy, West China Hospital, Sichuan University, Chengdu, China. hzheng@scu.edu.cn
- 2Laboratory of Pathology, West China Hospital, Sichuan University, Chengdu, China.
- 3Dizal (Jiangsu) Pharmaceutical Co., Ltd., Shanghai, China. hzheng@scu.edu.cn
- 4Cancer Center, West China Hospital, Sichuan University, Chengdu, China.
- 5Department of Epidemiology and Bio-Statistics, West China School of Public Health, Sichuan University, Chengdu, China.
- KMID: 2437606
- DOI: http://doi.org/10.4143/crt.2017.598
Abstract
- PURPOSE
The prevalence of PIK3CA in Chinese breast cancer patients may be underestimated. Therefore, we investigated the distribution of somatic PIK3CA/AKT1 mutations in Chinese breast cancer patients and explored their roles in tumor phenotypes and disease prognosis.
MATERIALS AND METHODS
Tumors from 507 breast cancer patients were prospectively collected from the West China Hospital between 2008 and 2013. Whole exons of AKT1 and PIK3CA were detected in fresh-frozen tumors using next-generation sequencing, and correlations between PIK3CA/AKT1 mutations and clinicopathological features were analyzed.
RESULTS
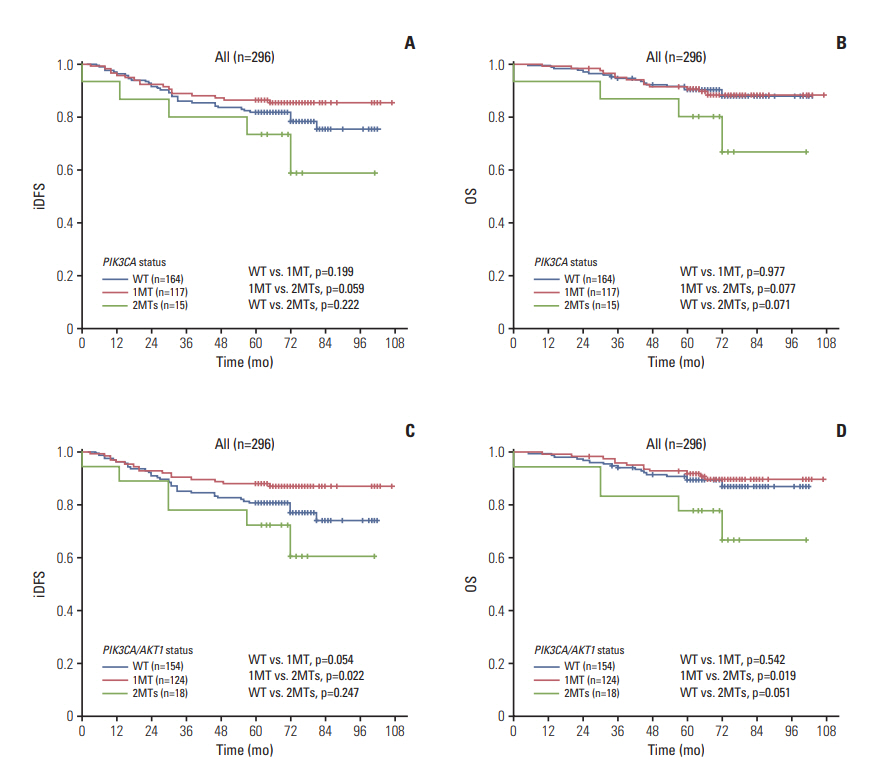
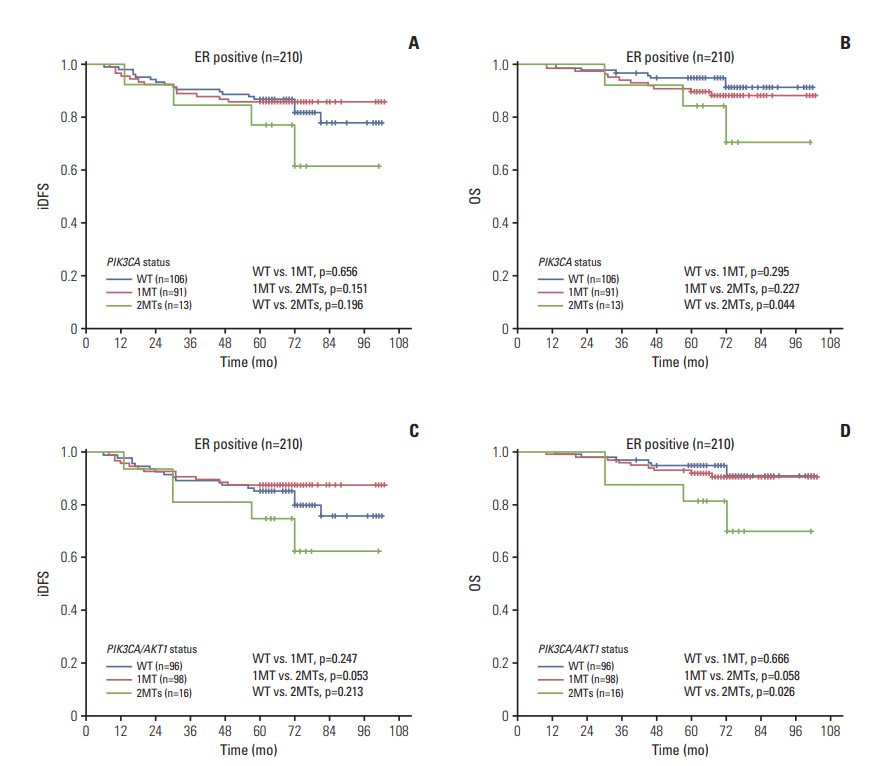
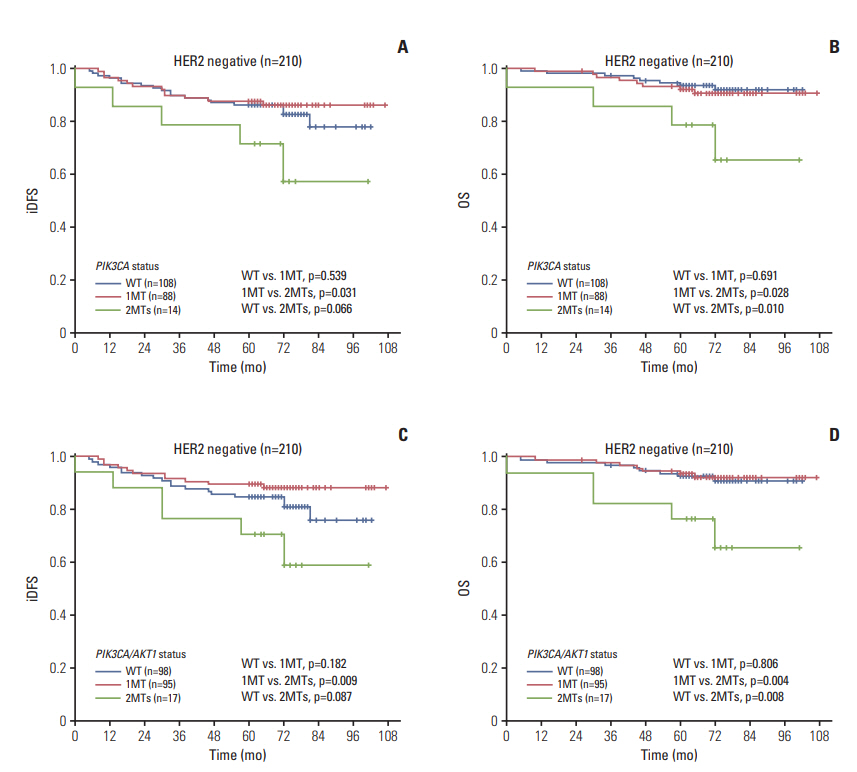
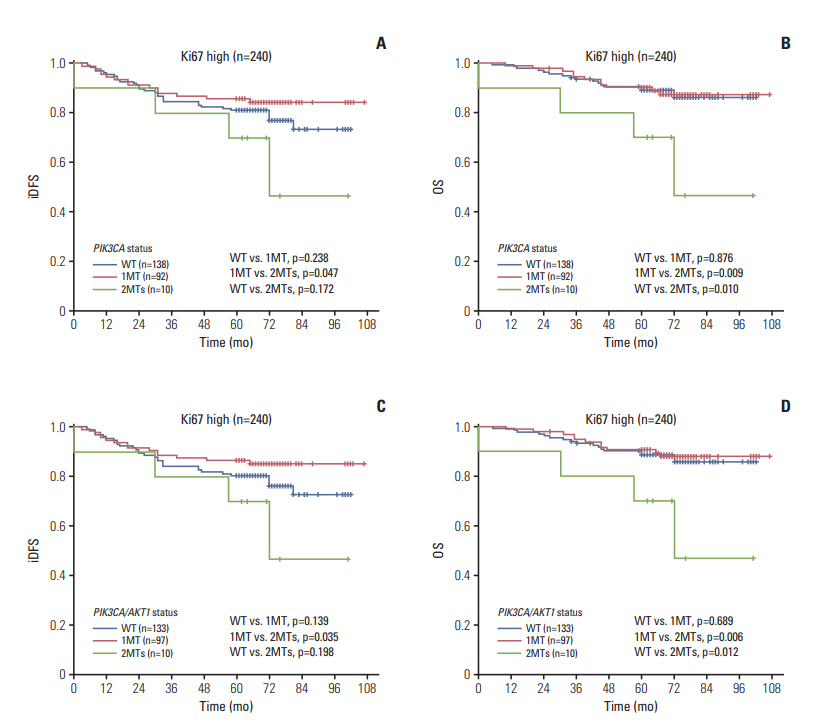
The AKT1 mutation was found in 3.6% (18/507) of patients. Tumors from patients that carried the AKT1 mutation were estrogen receptor (ER)+/progesterone receptor (PR)+/human epidermal growth factor receptor 2 (HER2)"’ and were more likely to have high expression levels of Ki67. The prevalence of the PIK3CA mutation was 46.5% (236/507), and 35 patients carried two or three variants of the PIK3CA gene. PIK3CA mutations were associated with ER+/PR+/HER2"’ status. The prognosis of patients with one mutation in PIK3CA (or PIK3CA/AKT1) was not significantly different than that of patients with wild-type PIK3CA (or PIK3CA/AKT1), while patients with two or three variants in PIK3CA (or PIK3CA/AKT1) exhibited poorer outcomes in the entire group and in all three subgroups (ER+, HER2"’, Ki67 high), particularly with respect to overall survival.
CONCLUSION
A high frequency of somatic PIK3CA mutations was detected in Chinese breast cancer patients. In addition to the mutation frequency, the tumor mutational burden of the PIK3CA and AKT1 genes should also be of concern, as they may be associated with poor prognosis.
Keyword
MeSH Terms
Figure
Cited by 2 articles
-
PIK3CA H1047R Mutation Associated with a Lower Pathological Complete Response Rate in Triple-Negative Breast Cancer Patients Treated with Anthracycline-Taxane–Based Neoadjuvant Chemotherapy
Sanxing Guo, Sibylle Loibl, Gunter von Minckwitz, Silvia Darb-Esfahani, Bianca Lederer, Carsten Denkert
Cancer Res Treat. 2020;52(3):689-696. doi: 10.4143/crt.2019.497.Analysis of
PIK3CA Mutation Concordance and Frequency in Primary and Different Distant Metastatic Sites in Breast Cancer
Jieun Park, Soo Youn Cho, Eun Sol Chang, Minjung Sung, Ji-Young Song, Kyungsoo Jung, Sung-Su Kim, Young Kee Shin, Yoon-La Choi
Cancer Res Treat. 2023;55(1):145-154. doi: 10.4143/crt.2022.001.
Reference
-
References
1. Rodon J, Dienstmann R, Serra V, Tabernero J. Development of PI3K inhibitors: lessons learned from early clinical trials. Nat Rev Clin Oncol. 2013; 10:143–53.
Article2. Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2015; 15:7–24.
Article3. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012; 490:61–70.4. Cizkova M, Dujaric ME, Lehmann-Che J, Scott V, Tembo O, Asselain B, et al. Outcome impact of PIK3CA mutations in HER2-positive breast cancer patients treated with trastuzumab. Br J Cancer. 2013; 108:1807–9.
Article5. Loi S, Michiels S, Lambrechts D, Fumagalli D, Claes B, Kellokumpu-Lehtinen PL, et al. Somatic mutation profiling and associations with prognosis and trastuzumab benefit in early breast cancer. J Natl Cancer Inst. 2013; 105:960–7.
Article6. Cizkova M, Vacher S, Meseure D, Trassard M, Susini A, Mlcuchova D, et al. PIK3R1 underexpression is an independent prognostic marker in breast cancer. BMC Cancer. 2013; 13:545.
Article7. Sabine VS, Crozier C, Brookes CL, Drake C, Piper T, van de Velde CJ, et al. Mutational analysis of PI3K/AKT signaling pathway in tamoxifen exemestane adjuvant multinational pathology study. J Clin Oncol. 2014; 32:2951–8.
Article8. Loibl S, von Minckwitz G, Schneeweiss A, Paepke S, Lehmann A, Rezai M, et al. PIK3CA mutations are associated with lower rates of pathologic complete response to anti-human epidermal growth factor receptor 2 (her2) therapy in primary HER2-overexpressing breast cancer. J Clin Oncol. 2014; 32:3212–20.
Article9. Majewski IJ, Nuciforo P, Mittempergher L, Bosma AJ, Eidtmann H, Holmes E, et al. PIK3CA mutations are associated with decreased benefit to neoadjuvant human epidermal growth factor receptor 2-targeted therapies in breast cancer. J Clin Oncol. 2015; 33:1334–9.
Article10. Deng L, Chen J, Zhong XR, Luo T, Wang YP, Huang HF, et al. Correlation between activation of PI3K/AKT/mTOR pathway and prognosis of breast cancer in Chinese women. PLoS One. 2015; 10:e0120511.
Article11. Yuan H, Chen J, Liu Y, Ouyang T, Li J, Wang T, et al. Association of PIK3CA mutation status before and after neoadjuvant chemotherapy with response to chemotherapy in women with breast cancer. Clin Cancer Res. 2015; 21:4365–72.
Article12. Papaxoinis G, Kotoula V, Alexopoulou Z, Kalogeras KT, Zagouri F, Timotheadou E, et al. Significance of PIK3CA mutations in patients with early breast cancer treated with adjuvant chemotherapy: a Hellenic Cooperative Oncology Group (HeCOG) Study. PLoS One. 2015; 10:e0140293.
Article13. Engels CC, Kiderlen M, Bastiaannet E, van Eijk R, Mooyaart A, Smit VT, et al. The clinical value of HER-2 overexpression and PIK3CA mutations in the older breast cancer population: a FOCUS study analysis. Breast Cancer Res Treat. 2016; 156:361–70.
Article14. Xu B, Guan Z, Shen Z, Tong Z, Jiang Z, Yang J, et al. Association of phosphatase and tensin homolog low and phosphatidylinositol 3-kinase catalytic subunit alpha gene mutations on outcome in human epidermal growth factor receptor 2-positive metastatic breast cancer patients treated with first-line lapatinib plus paclitaxel or paclitaxel alone. Breast Cancer Res. 2014; 16:405.
Article15. Hortobagyi GN, Chen D, Piccart M, Rugo HS, Burris HA 3rd, Pritchard KI, et al. Correlative analysis of genetic alterations and everolimus benefit in hormone receptor-positive, human epidermal growth factor receptor 2-negative advanced breast cancer: Results From BOLERO-2. J Clin Oncol. 2016; 34:419–26.
Article16. Pang B, Cheng S, Sun SP, An C, Liu ZY, Feng X, et al. Prognostic role of PIK3CA mutations and their association with hormone receptor expression in breast cancer: a meta-analysis. Sci Rep. 2014; 4:6255.
Article17. Guideline Recommendations for Immunohistochemistry Detection in Breast Cancer Group. Guideline for testing of estrogen and progesterone receptors in breast cancer. Zhonghua Bing Li Xue Za Zhi. 2015; 44:237–9.18. Guideline Recommendations for HER2 Detection in Breast Cancer Group. Guidelines for HER2 detection in breast cancer, the 2014 version. Zhonghua Bing Li Xue Za Zhi. 2014; 43:262–7.19. Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thurlimann B, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013; 24:2206–23.20. Gourgou-Bourgade S, Cameron D, Poortmans P, Asselain B, Azria D, Cardoso F, et al. Guidelines for time-to-event end point definitions in breast cancer trials: results of the DATECAN initiative (Definition for the Assessment of Time-toevent Endpoints in CANcer trials). Ann Oncol. 2015; 26:2505–6.21. Ahmad F, Badwe A, Verma G, Bhatia S, Das BR. Molecular evaluation of PIK3CA gene mutation in breast cancer: determination of frequency, distribution pattern and its association with clinicopathological findings in Indian patients. Med Oncol. 2016; 33:74.
Article22. Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, Inao T, Sueta A, Fujiwara S, et al. Prognostic role of PIK3CA mutations of cell-free DNA in early-stage triple negative breast cancer. Cancer Sci. 2015; 106:1582–9.23. Leo F, Bartels S, Magel L, Framke T, Busche G, Jonigk D, et al. Prognostic factors in the myoepithelial-like spindle cell type of metaplastic breast cancer. Virchows Arch. 2016; 469:191–201.
Article24. Jacot W, Mollevi C, Fina F, Lopez-Crapez E, Martin PM, Colombo PE, et al. High EGFR protein expression and exon 9 PIK3CA mutations are independent prognostic factors in triple negative breast cancers. BMC Cancer. 2015; 15:986.
Article25. Baselga J, Cortes J, Im SA, Clark E, Ross G, Kiermaier A, et al. Biomarker analyses in CLEOPATRA: a phase III, placebo-controlled study of pertuzumab in human epidermal growth factor receptor 2-positive, first-line metastatic breast cancer. J Clin Oncol. 2014; 32:3753–61.
Article26. Kim JY, Lee E, Park K, Park WY, Jung HH, Ahn JS, et al. Clinical implications of genomic profiles in metastatic breast cancer with a focus on TP53 and PIK3CA, the most frequently mutated genes. Oncotarget. 2017; 8:27997–8007.
Article27. Jensen JD, Knoop A, Laenkholm AV, Grauslund M, Jensen MB, Santoni-Rugiu E, et al. PIK3CA mutations, PTEN, and pHER2 expression and impact on outcome in HER2-positive early-stage breast cancer patients treated with adjuvant chemotherapy and trastuzumab. Ann Oncol. 2012; 23:2034–42.
Article28. Ibrahim EM, Kazkaz GA, Al-Mansour MM, Al-Foheidi ME. The predictive and prognostic role of phosphatase phosphoinositol-3 (PI3) kinase (PIK3CA) mutation in HER2-positive breast cancer receiving HER2-targeted therapy: a meta-analysis. Breast Cancer Res Treat. 2015; 152:463–76.
Article29. Loibl S, Majewski I, Guarneri V, Nekljudova V, Holmes E, Bria E, et al. PIK3CA mutations are associated with reduced pathological complete response rates in primary HER2-positive breast cancer: pooled analysis of 967 patients from five prospective trials investigating lapatinib and trastuzumab. Ann Oncol. 2016; 27:1519–25.
Article30. Yang SX, Polley E, Lipkowitz S. New insights on PI3K/AKT pathway alterations and clinical outcomes in breast cancer. Cancer Treat Rev. 2016; 45:87–96.
Article31. Oda K, Okada J, Timmerman L, Rodriguez-Viciana P, Stokoe D, Shoji K, et al. PIK3CA cooperates with other phosphatidylinositol 3'-kinase pathway mutations to effect oncogenic transformation. Cancer Res. 2008; 68:8127–36.
Article32. Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo WL, Davies M, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008; 68:6084–91.
Article33. Moynahan ME, Chen D, He W, Sung P, Samoila A, You D, et al. Correlation between PIK3CA mutations in cell-free DNA and everolimus efficacy in HR(+), HER2(–) advanced breast cancer: results from BOLERO-2. Br J Cancer. 2017; 116:726–30.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Concomitant PIK3CA and TP53 Mutations in Breast Cancer: An Analysis of Clinicopathologic and Mutational Features, Neoadjuvant Therapeutic Response, and Prognosis
- Somatic Mutations of TP53 Identified by Targeted Next-Generation Sequencing Are Poor Prognostic Factors for Primary Operable Breast Cancer: A Single-Center Study
- The Significance of p-AKT1 as a Prognostic Marker and Therapeutic Target in Patients With Hormone Receptor-Positive and Human Epidermal Growth Factor Receptor-2-Positive Early Breast Cancer
- Biomarkers of Everolimus Sensitivity in Hormone Receptor-Positive Breast Cancer
- PIK3CA Mutation is Associated with Poor Response to HER2-Targeted Therapy in Breast Cancer Patients