Ann Lab Med.
2012 Nov;32(6):445-449.
Additional Genomic Aberrations Identified by Single Nucleotide Polymorphism Array-Based Karyotyping in an Acute Myeloid Leukemia Case with Isolated del(20q) Abnormality
- Affiliations
-
- 1Department of Laboratory Medicine, Ewha Womans University School of Medicine, Seoul, Korea. JungWonH@ewha.ac.kr
- 2Department of Internal Medicine, Ewha Womans University School of Medicine, Seoul, Korea.
Abstract
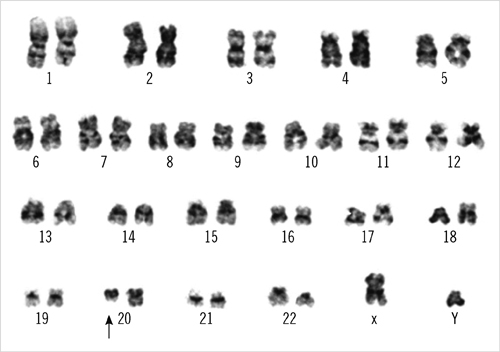
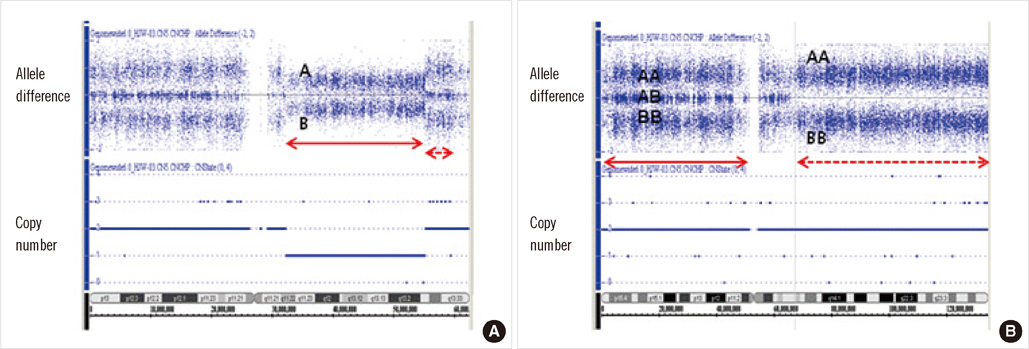
- Prognosis is known to be better in cases with isolated chromosomal abnormalities than in those with complex karyotypes. Accordingly, del(20q) as an isolated abnormality must be distinguished from cases in which it is associated with other chromosomal rearrangements for a better stratification of prognosis. We report a case of an isolated del(20q) abnormality with additional genomic aberrations identified using whole-genome single nucleotide polymorphism array (SNP-A)-based karyotyping. A 39-yr-old man was diagnosed with AML without maturation. Metaphase cytogenetic analysis (MC) revealed del(20)(q11.2) as the isolated abnormality in 100% of metaphase cells analyzed, and FISH analysis using D20S108 confirmed the 20q deletion in 99% of interphase cells. Using FISH, other rearrangements such as BCR/ABL1, RUNX1/RUNX1T1, PML/RARA, CBFB/MYH11, and MLL were found to be negative. SNP-A identified an additional copy neutral loss of heterozygosity (CN-LOH) in the 11q13.1-q25 region. Furthermore, SNP-A allowed for a more precise definition of the breakpoints of the 20q deletion (20q11.22-q13.31). Unexpectedly, the terminal regions showed gain on chromosome 20q. The patient did not achieve complete remission; 8 months later, he died from complications of leukemic cell infiltrations into the central nervous system. This study suggests that a presumably isolated chromosomal abnormality by MC may have additional genomic aberrations, including CN-LOH, which could be associated with a poor prognosis. SNP-A-based karyotyping may be helpful for distinguishing true isolated cases from cases in combination with additional genomic aberrations not detected by MC.
Keyword
MeSH Terms
-
Adult
Antineoplastic Agents/therapeutic use
Chromosome Deletion
*Chromosomes, Human, Pair 20
Cytogenetic Analysis
Humans
Karyotyping
Leukemia, Myeloid, Acute/*diagnosis/drug therapy/genetics
Loss of Heterozygosity
Male
Oligonucleotide Array Sequence Analysis
Polymorphism, Single Nucleotide
Antineoplastic Agents
Figure
Reference
-
1. Atlas of genetics and cytogenetics in oncology and hematology. Updated on Oct 2011. http://atlasgeneticsoncology.org/Anomalies/del20qID1040.html.2. Swerdlow SH, Campo E, editors. WHO classification of tumours of haematopoietic and lymphoid tissues. 2008. 4th ed. Lyon: IARC.3. Brezinová J, Zemanová Z, Ransdorfová S, Sindelárová L, Sisková M, Neuwirtová R, et al. Prognostic significance of del(20q) in patients with hematological malignancies. Cancer Genet Cytogenet. 2005. 160:188–192.4. Maciejewski JP, Tiu RV, O'Keefe C. Application of array-based whole genome scanning technologies as a cytogenetic tool in haematological malignancies. Br J Haematol. 2009. 146:479–488.
Article5. Makishima H, Maciejewski JP. Pathogenesis and consequences of uniparental disomy in cancer. Clin Cancer Res. 2011. 17:3913–3923.
Article6. Huh J, Tiu RV, Gondek LP, O'Keefe CL, Jasek M, Makishima H, et al. Characterization of chromosome arm 20q abnormalities in myeloid malignancies using genome-wide single nucleotide polymorphism array analysis. Genes Chromosomes Cancer. 2010. 49:390–399.
Article7. Barresi V, Palumbo GA, Musso N, Consoli C, Capizzi C, Meli CR, et al. Clonal selection of 11q CN-LOH and CBL gene mutation in a serially studied patient during MDS progression to AML. Leuk Res. 2010. 34:1539–1542.
Article8. Dunbar AJ, Gondek LP, O'Keefe CL, Makishima H, Rataul MS, Szpurka H, et al. 250K single nucleotide polymorphism array karyotyping identifies acquired uniparental disomy and homozygous mutation, including novel missense substitutions of c-Cbl, in myeloid malignancies. Cancer Res. 2008. 68:10349–10357.9. Grand FH, Hidalgo-Curtis CE, Ernst T, Zoi K, Zoi C, McGuire C, et al. Frequent CBL mutations associated with 11q acquired uniparental disomy in myeloproliferative neoplasms. Blood. 2009. 113:6182–6192.
Article10. Evers C, Beier M, Poelitz A, Hildebrandt B, Servan K, Drechsler M, et al. Molecular definition of chromosome arm 5q deletion end points and detection of hidden aberrations in patients with myelodysplastic syndromes and isolated del(5q) using oligonucleotide array CGH. Genes Chromosomes Cancer. 2007. 46:1119–1128.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Ring Chromosome 5 in Acute Myeloid Leukemia Defined by Whole-genome Single Nucleotide Polymorphism Array
- A Case of Acute Myeloid Leukemia with Bone Marrow Basophilia and Dysmegakaryocytic Hyperplasia with Isochromosome 17q as a Sole Cytogenetic Abnormality: A Clinical Study with Literature Review
- Clinical Significance of Previously Cryptic Copy Number Alterations and Loss of Heterozygosity in Pediatric Acute Myeloid Leukemia and Myelodysplastic Syndrome Determined Using Combined Array Comparative Genomic Hybridization plus Single-Nucleotide Polymorphism Microarray Analyses
- A Case of Near-triploidy in Myelodysplastic Syndrome with del(5q) Combined with del(1p) and del(13q)
- Chromothripsis Identified by Copy Number Profiling in a Case of Plasma Cell Leukaemia