J Breast Cancer.
2018 Dec;21(4):363-370. 10.4048/jbc.2018.21.e56.
Identification and Validation of Circulating MicroRNA Signatures for Breast Cancer Early Detection Based on Large Scale Tissue-Derived Data
- Affiliations
-
- 1School of Biology and Biological Engineering, South China University of Technology, Guangzhou, China. hldu@scut.edu.cn
- 2Department of Laboratory Science, The Second Affiliated Hospital of Guangzhou University of Chinese Medicine, Guangzhou, China.
- 3Department of Laboratory Medicine, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Sun Yat-sen University Cancer Center, Guangzhou, China.
- KMID: 2429814
- DOI: http://doi.org/10.4048/jbc.2018.21.e56
Abstract
- PURPOSE
Breast cancer is the most commonly occurring cancer among women worldwide, and therefore, improved approaches for its early detection are urgently needed. As microRNAs (miRNAs) are increasingly recognized as critical regulators in tumorigenesis and possess excellent stability in plasma, this study focused on using miRNAs to develop a method for identifying noninvasive biomarkers.
METHODS
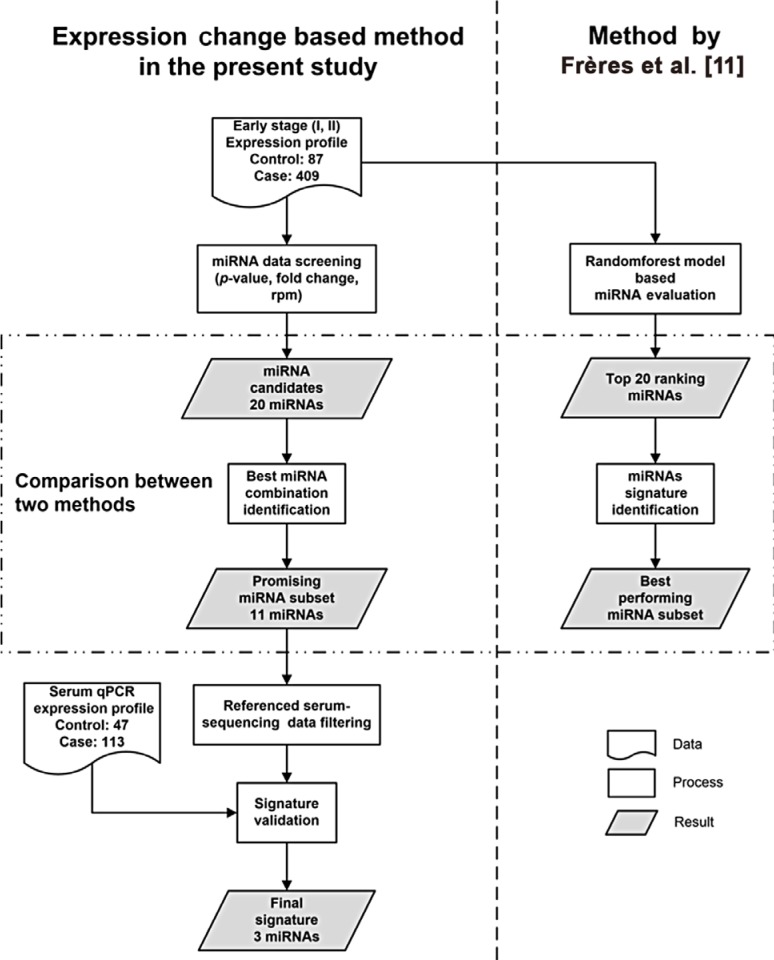
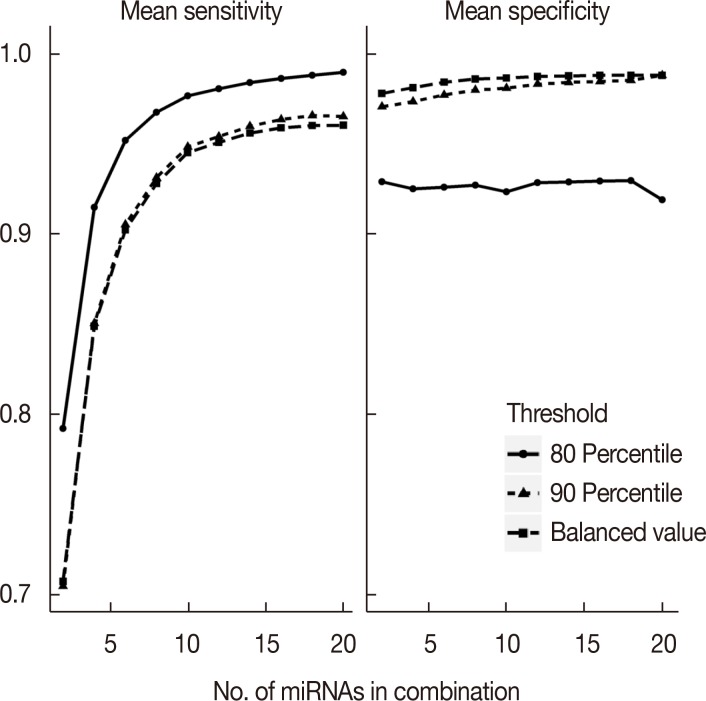
To discover critical candidates, differential expression analysis was performed on tissue-originated miRNA profiles of 409 early breast cancer patients and 87 healthy controls from The Cancer Genome Atlas database. We selected candidates from the differentially expressed miRNAs and then evaluated every possible molecular signature formed by the candidates. The best signature was validated in independent serum samples from 113 early breast cancer patients and 47 healthy controls using reverse transcription quantitative real-time polymerase chain reaction.
RESULTS
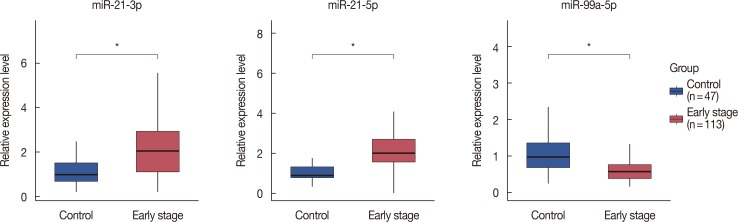
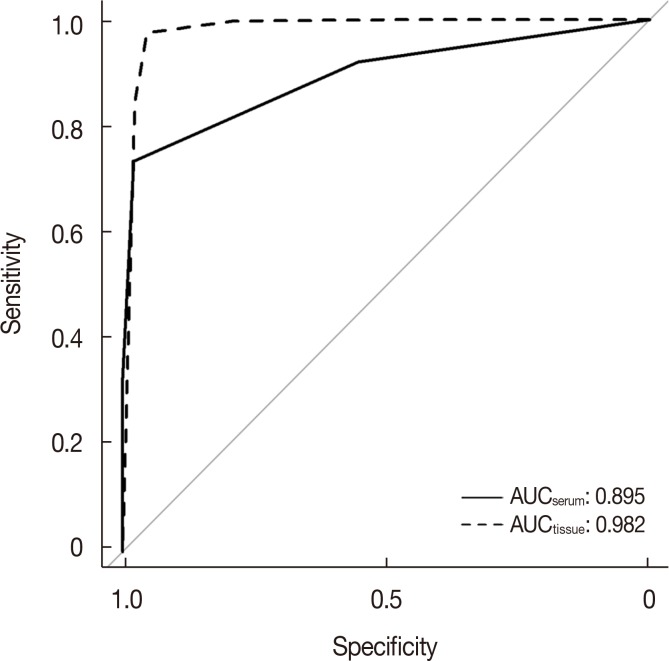
The miRNA candidates in our method were revealed to be associated with breast cancer according to previous studies and showed potential as useful biomarkers. When validated in independent serum samples, the area under curve of the final miRNA signature (miR-21-3p, miR-21-5p, and miR-99a-5p) was 0.895. Diagnostic sensitivity and specificity were 97.9% and 73.5%, respectively.
CONCLUSION
The present study established a novel and effective method to identify biomarkers for early breast cancer. And the method, is also suitable for other cancer types. Furthermore, a combination of three miRNAs was identified as a prospective biomarker for breast cancer early detection.
MeSH Terms
Figure
Reference
-
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015; 65:87–108. PMID: 25651787.
Article2. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Oncol Pract. 2007; 3:336–339. PMID: 29436954.3. Lin XJ, Chong Y, Guo ZW, Xie C, Yang XJ, Zhang Q, et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: a multicentre, retrospective, longitudinal biomarker identification study with a nested case-control study. Lancet Oncol. 2015; 16:804–815. PMID: 26088272.
Article4. Du M, Shi D, Yuan L, Li P, Chu H, Qin C, et al. Circulating miR-497 and miR-663b in plasma are potential novel biomarkers for bladder cancer. Sci Rep. 2015; 5:10437. PMID: 26014226.
Article5. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008; 105:10513–10518. PMID: 18663219.
Article6. Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008; 18:997–1006. PMID: 18766170.
Article7. Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008; 3:e3148. PMID: 18773077.
Article8. Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010; 38:7248–7259. PMID: 20615901.
Article9. Pigati L, Yaddanapudi SC, Iyengar R, Kim DJ, Hearn SA, Danforth D, et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One. 2010; 5:e13515. PMID: 20976003.
Article10. Kayala MA, Baldi P. Cyber-T web server: differential analysis of high-throughput data. Nucleic Acids Res. 2012; 40:W553–W559. PMID: 22600740.
Article11. Frères P, Wenric S, Boukerroucha M, Fasquelle C, Thiry J, Bovy N, et al. Circulating microRNA-based screening tool for breast cancer. Oncotarget. 2016; 7:5416–5428. PMID: 26734993.
Article12. Allaya N, Khabir A, Sallemi-Boudawara T, Sellami N, Daoud J, Ghorbel A, et al. Over-expression of miR-10b in NPC patients: correlation with LMP1 and Twist1. Tumour Biol. 2015; 36:3807–3814. PMID: 25597482.
Article13. Chang YY, Kuo WH, Hung JH, Lee CY, Lee YH, Chang YC, et al. Deregulated microRNAs in triple-negative breast cancer revealed by deep sequencing. Mol Cancer. 2015; 14:36. PMID: 25888956.
Article14. Song C, Zhang L, Wang J, Huang Z, Li X, Wu M, et al. High expression of microRNA-183/182/96 cluster as a prognostic biomarker for breast cancer. Sci Rep. 2016; 6:24502. PMID: 27071841.
Article15. Calvano Filho CM, Calvano-Mendes DC, Carvalho KC, Maciel GA, Ricci MD, Torres AP, et al. Triple-negative and luminal A breast tumors: differential expression of miR-18a-5p, miR-17-5p, and miR-20a-5p. Tumour Biol. 2014; 35:7733–7741. PMID: 24810926.
Article16. Matamala N, Vargas MT, González-Cámpora R, Miñambres R, Arias JI, Menéndez P, et al. Tumor microRNA expression profiling identifies circulating microRNAs for early breast cancer detection. Clin Chem. 2015; 61:1098–1106. PMID: 26056355.
Article17. Ouyang M, Li Y, Ye S, Ma J, Lu L, Lv W, et al. MicroRNA profiling implies new markers of chemoresistance of triple-negative breast cancer. PLoS One. 2014; 9:e96228. PMID: 24788655.
Article18. Mar-Aguilar F, Mendoza-Ramírez JA, Malagón-Santiago I, Espino-Silva PK, Santuario-Facio SK, Ruiz-Flores P, et al. Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis Markers. 2013; 34:163–169. PMID: 23334650.
Article19. Li J, Song ZJ, Wang YY, Yin Y, Liu Y, Nan X. Low levels of serum miR-99a is a predictor of poor prognosis in breast cancer. Genet Mol Res. 2016; 15(3):gmr8338.
Article20. Pan Y, Zhang J, Fu H, Shen L. miR-144 functions as a tumor suppressor in breast cancer through inhibiting ZEB1/2-mediated epithelial mesenchymal transition process. Onco Targets Ther. 2016; 9:6247–6255. PMID: 27785072.
Article21. Madhavan D, Peng C, Wallwiener M, Zucknick M, Nees J, Schott S, et al. Circulating miRNAs with prognostic value in metastatic breast cancer and for early detection of metastasis. Carcinogenesis. 2016; 37:461–470. PMID: 26785733.
Article22. Zhang G, Liu Z, Cui G, Wang X, Yang Z. MicroRNA-486-5p targeting PIM-1 suppresses cell proliferation in breast cancer cells. Tumour Biol. 2014; 35:11137–11145. PMID: 25104088.
Article23. Zhu J, Zheng Z, Wang J, Sun J, Wang P, Cheng X, et al. Different miRNA expression profiles between human breast cancer tumors and serum. Front Genet. 2014; 5:149. PMID: 24904649.
Article24. Si H, Sun X, Chen Y, Cao Y, Chen S, Wang H, et al. Circulating microRNA-92a and microRNA-21 as novel minimally invasive biomarkers for primary breast cancer. J Cancer Res Clin Oncol. 2013; 139:223–229. PMID: 23052693.
Article25. Kodahl AR, Lyng MB, Binder H, Cold S, Gravgaard K, Knoop AS, et al. Novel circulating microRNA signature as a potential non-invasive multi-marker test in ER-positive early-stage breast cancer: a case control study. Mol Oncol. 2014; 8:874–883. PMID: 24694649.
Article26. Shimomura A, Shiino S, Kawauchi J, Takizawa S, Sakamoto H, Matsuzaki J, et al. Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer Sci. 2016; 107:326–334. PMID: 26749252.
Article27. Dai X, Fang M, Li S, Yan Y, Zhong Y, Du B. miR-21 is involved in transforming growth factor beta1-induced chemoresistance and invasion by targeting PTEN in breast cancer. Oncol Lett. 2017; 14:6929–6936. PMID: 29151919.28. Doberstein K, Bretz NP, Schirmer U, Fiegl H, Blaheta R, Breunig C, et al. miR-21-3p is a positive regulator of L1CAM in several human carcinomas. Cancer Lett. 2014; 354:455–466. PMID: 25149066.
Article29. Xia M, Li H, Wang JJ, Zeng HJ, Wang SH. MiR-99a suppress proliferation, migration and invasion through regulating insulin-like growth factor 1 receptor in breast cancer. Eur Rev Med Pharmacol Sci. 2016; 20:1755–1763. PMID: 27212167.30. Fan T, Mao Y, Sun Q, Liu F, Lin JS, Liu Y, et al. Branched rolling circle amplification method for measuring serum circulating microRNA levels for early breast cancer detection. Cancer Sci. 2018; 109:2897–2906. PMID: 29981251.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Evaluation of the Value of Multiplex MicroRNA Analysis as a Breast Cancer Screening in Korean Women under 50 Years of Age with a High Proportion of Dense Breasts
- Circulating Tumor Cells: Detection Methods and Potential Clinical Application in Breast Cancer
- Circulating Tumor Cells and Extracellular Nucleic Acids in Breast Cancer
- Circulating Cell-free Tumor Nucleic Acids in Gastric Cancer
- Application of Artificial Intelligence in Breast Imaging: Current Landscape and Prospects