Blood Res.
2018 Sep;53(3):240-249. 10.5045/br.2018.53.3.240.
Spectrum of mitochondrial genome instability and implication of mitochondrial haplogroups in Korean patients with acute myeloid leukemia
- Affiliations
-
- 1College of Korean Medicine, Dongshin University, Naju, Korea.
- 2Department of Laboratory Medicine, Gwangyang Sarang General Hospital, Gwangyang, Korea.
- 3Department of Laboratory Medicine, Chonnam National University Medical School and Chonnam National University Hwasun Hospital, Hwasun, Korea. mgshin@chonnam.ac.kr
- 4Brain Korea 21 Plus Project, Chonnam National University Medical School, Gwangju, Korea.
- 5Environmental Health Center for Childhood Leukemia and Cancer, Chonnam National University Medical School and Chonnam National University Hwasun Hospital, Hwasun, Korea.
- KMID: 2429325
- DOI: http://doi.org/10.5045/br.2018.53.3.240
Abstract
- BACKGROUND
Mitochondrial DNA (mtDNA) mutations may regulate the progression and chemosensitivity of leukemia. Few studies regarding mitochondrial aberrations and haplogroups in acute myeloid leukemia (AML) and their clinical impacts have been reported. Therefore, we focused on the mtDNA length heteroplasmies minisatellite instability (MSI), copy number alterations, and distribution of mitochondrial haplogroups in Korean patients with AML.
METHODS
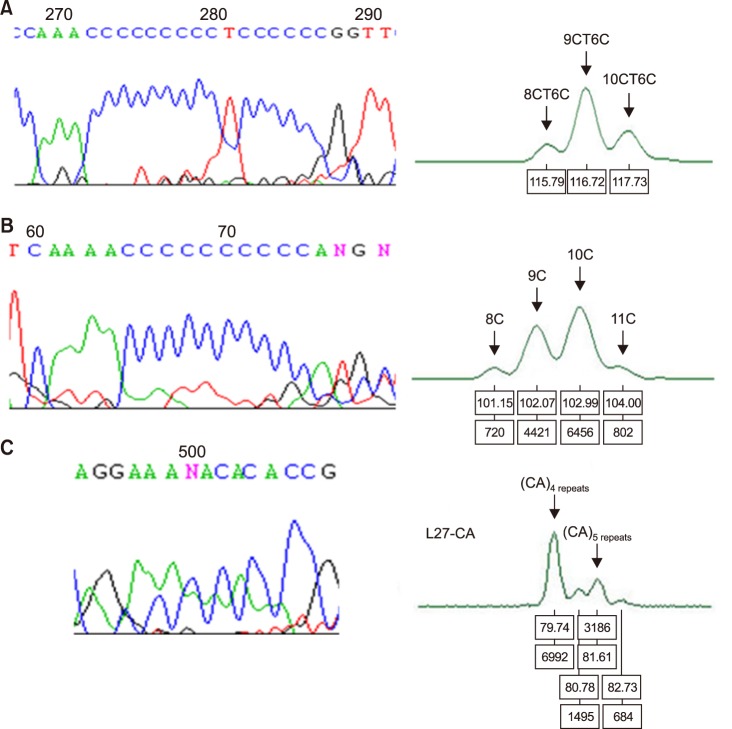
This study investigated 74 adult patients with AML and 70 controls to evaluate mtDNA sequence alterations, MSI, mtDNA copy number, haplogroups, and their clinical implications. The hypervariable (HV) control regions (HV1 and HV2), tRNA(leu1)gene, and cytochrome b gene of mtDNA were analyzed. Two mtDNA minisatellite markers, 16189 poly-C (¹â¶¹â¸â´CCCCCTCCCC¹â¶¹â¹³, 5CT4C) and 303 poly-C (³â°³CCCCCCCTCCCCC³¹âµ, 7CT5C), were used to examine the mtDNA MSI.
RESULTS
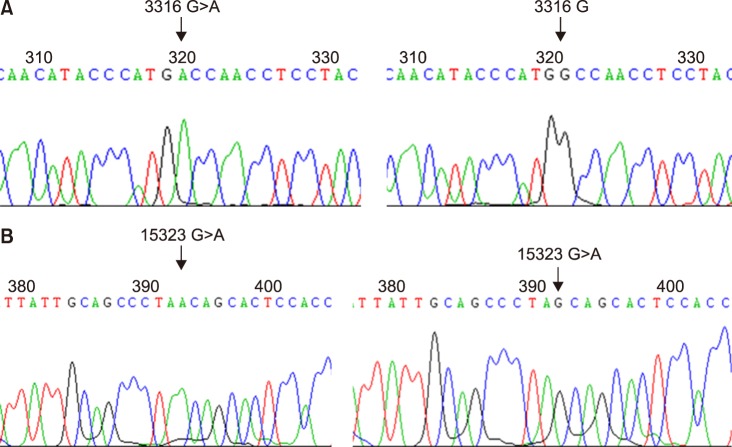
In AML, most mtDNA sequence variants were single nucleotide substitutions, but there were no significant differences compared to those in controls. The number of mtMSI patterns increased in AML. The mean mtDNA copy number of AML patients increased approximately 9-fold compared to that of controls (P < 0.0001). Haplogroup D4 was found in AML with a higher frequency compared to that in controls (31.0% vs. 15.7%, P=0.046). None of the aforementioned factors showed significant impacts on the outcomes.
CONCLUSION
AML cells disclosed more heterogeneous patterns with the mtMSI markers and had increased mtDNA copy numbers. These findings implicate mitochondrial genome instability in primary AML cells. Therefore, mtDNA haplogroup D4 might be associated with AML risk among Koreans.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Comparative evaluation of the developed targeted RNA sequencing system and a commercialized test panel
Young Eun Lee, Ju Heon Park, Ha Jin Lim, Hye Ran Kim, Jong Hee Shin, Myung Geun Shin
Blood Res. 2022;57(3):235-238. doi: 10.5045/br.2022.2022095.
Reference
-
1. Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997; 66:409–435. PMID: 9242913.
Article2. DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N Engl J Med. 2003; 348:2656–2668. PMID: 12826641.
Article3. Anderson S, Bankier AT, Barrell BG, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981; 290:457–465. PMID: 7219534.
Article4. Penta JS, Johnson FM, Wachsman JT, Copeland WC. Mitochondrial DNA in human malignancy. Mutat Res. 2001; 488:119–133. PMID: 11344040.
Article5. Shin MG, Kajigaya S, McCoy JP Jr, Levin BC, Young NS. Marked mitochondrial DNA sequence heterogeneity in single CD34+ cell clones from normal adult bone marrow. Blood. 2004; 103:553–561. PMID: 14504082.
Article6. Shin MG, Levin BC, Kim HJ, et al. Profiling of length heteroplasmies in the human mitochondrial DNA control regions from blood cells in the Korean population. Electrophoresis. 2006; 27:1331–1340. PMID: 16502464.
Article7. Park SY, Shin MG, Kim HR, et al. Alteration of mitochondrial DNA sequence and copy number in nasal polyp tissue. Mitochondrion. 2009; 9:318–325. PMID: 19426839.
Article8. Lim SW, Kim HR, Kim HY, et al. High-frequency minisatellite instability of the mitochondrial genome in colorectal cancer tissue associated with clinicopathological values. Int J Cancer. 2012; 131:1332–1341. PMID: 22120612.
Article9. Veltri KL, Espiritu M, Singh G. Distinct genomic copy number in mitochondria of different mammalian organs. J Cell Physiol. 1990; 143:160–164. PMID: 2318903.
Article10. Shin MG, Kajigaya S, Levin BC, Young NS. Mitochondrial DNA mutations in patients with myelodysplastic syndromes. Blood. 2003; 101:3118–3125. PMID: 12446454.
Article11. Verma M, Naviaux RK, Tanaka M, Kumar D, Franceschi C, Singh KK. Meeting report: mitochondrial DNA and cancer epidemiology. Cancer Res. 2007; 67:437–439. PMID: 17213255.
Article12. Mondal R, Ghosh SK, Choudhury JH, et al. Mitochondrial DNA copy number and risk of oral cancer: a report from Northeast India. PLoS One. 2013; 8:e57771. PMID: 23469236.
Article13. Pakendorf B, Stoneking M. Mitochondrial DNA and human evolution. Annu Rev Genomics Hum Genet. 2005; 6:165–183. PMID: 16124858.
Article14. Torroni A, Huoponen K, Francalacci P, et al. Classification of European mtDNAs from an analysis of three European populations. Genetics. 1996; 144:1835–1850. PMID: 8978068.
Article15. Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005; 6:389–402. PMID: 15861210.
Article16. Shen L, Fang H, Chen T, et al. Evaluating mitochondrial DNA in cancer occurrence and development. Ann N Y Acad Sci. 2010; 1201:26–33. PMID: 20649535.
Article17. Estey EH. Acute myeloid leukemia: 2013 update on risk-stratification and management. Am J Hematol. 2013; 88:318–327. PMID: 23526416.
Article18. Carew JS, Zhou Y, Albitar M, Carew JD, Keating MJ, Huang P. Mitochondrial DNA mutations in primary leukemia cells after chemotherapy: clinical significance and therapeutic implications. Leukemia. 2003; 17:1437–1447. PMID: 12886229.
Article19. Grist SA, Lu XJ, Morley AA. Mitochondrial mutations in acute leukaemia. Leukemia. 2004; 18:1313–1316. PMID: 15129223.
Article20. Linnartz B, Anglmayer R, Zanssen S. Comprehensive scanning of somatic mitochondrial DNA alterations in acute leukemia developing from myelodysplastic syndromes. Cancer Res. 2004; 64:1966–1971. PMID: 15026331.
Article21. Sharawat SK, Bakhshi R, Vishnubhatla S, Bakhshi S. Mitochondrial D-loop variations in paediatric acute myeloid leukaemia: a potential prognostic marker. Br J Haematol. 2010; 149:391–398. PMID: 20230407.
Article22. Silkjaer T, Nørgaard JM, Aggerholm A, et al. Characterization and prognostic significance of mitochondrial DNA variations in acute myeloid leukemia. Eur J Haematol. 2013; 90:385–396. PMID: 23444869.
Article23. Shin MG, Kim HJ, Kim HR, et al. Mitochondrial DNA minisatellites as new markers for the quantitative determination of hematopoietic chimerism after allogeneic stem cell transplantation. Leukemia. 2007; 21:369–373. PMID: 17251903.
Article24. Kivisild T, Tolk HV, Parik J, et al. The emerging limbs and twigs of the East Asian mtDNA tree. Mol Biol Evol. 2002; 19:1737–1751. PMID: 12270900.
Article25. Yao YG, Kong QP, Bandelt HJ, Kivisild T, Zhang YP. Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am J Hum Genet. 2002; 70:635–651. PMID: 11836649.
Article26. He L, Luo L, Proctor SJ, et al. Somatic mitochondrial DNA mutations in adult-onset leukaemia. Leukemia. 2003; 17:2487–2491. PMID: 14523470.
Article27. Yao YG, Ogasawara Y, Kajigaya S, et al. Mitochondrial DNA sequence variation in single cells from leukemia patients. Blood. 2007; 109:756–762. PMID: 16946307.
Article28. Han H, Li DQ, Chen P, et al. Mutation detection of mitochondrial DNA D-loop region in bone marrow cells of acute leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2013; 21:29–33. PMID: 23484686.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Analysis of Nuclear Mitochondrial DNA Segments of Nine Plant Species: Size, Distribution, and Insertion Loci
- Genetics of Mitochondrial Myopathies
- MitGEN: Single Nucleotide Polymorphism DB Browser for Human Mitochondrial Genome
- Whole Mitochondrial Genome Sequence of an Indian Plasmodium falciparum Field Isolate
- Mechanisms of Uniparental Mitochondrial DNA Inheritance in Cryptococcus neoformans