J Vet Sci.
2018 Nov;19(6):759-770. 10.4142/jvs.2018.19.6.759.
Hexon and fiber gene changes in an attenuated fowl adenovirus isolate from Malaysia in embryonated chicken eggs and its infectivity in chickens
- Affiliations
-
- 1Faculty of Veterinary Medicine, Universiti Putra Malaysia, 43400 UPM Serdang, Malaysia. mdhair@upm.edu.my
- 2Institute of Bioscience, Universiti Putra Malaysia, 43400 UPM Serdang, Malaysia.
- 3Faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia, 43400 UPM Serdang, Malaysia.
- KMID: 2427024
- DOI: http://doi.org/10.4142/jvs.2018.19.6.759
Abstract
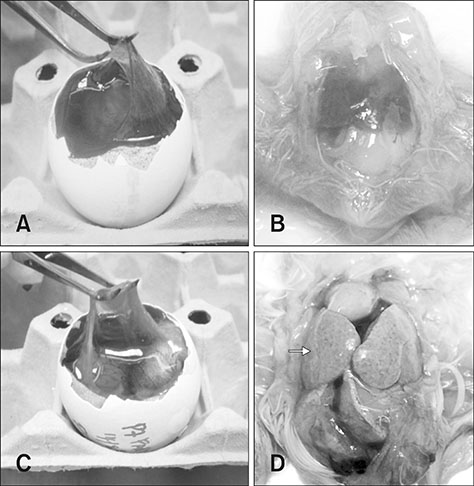
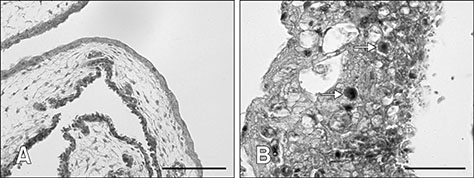
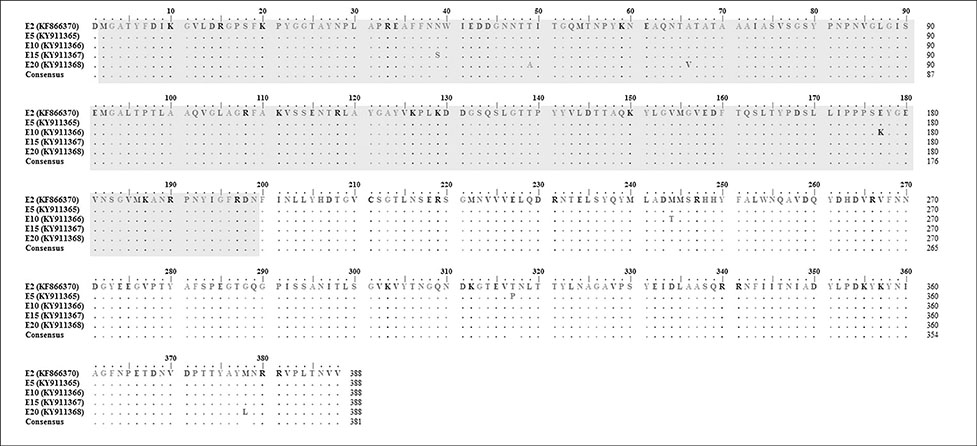
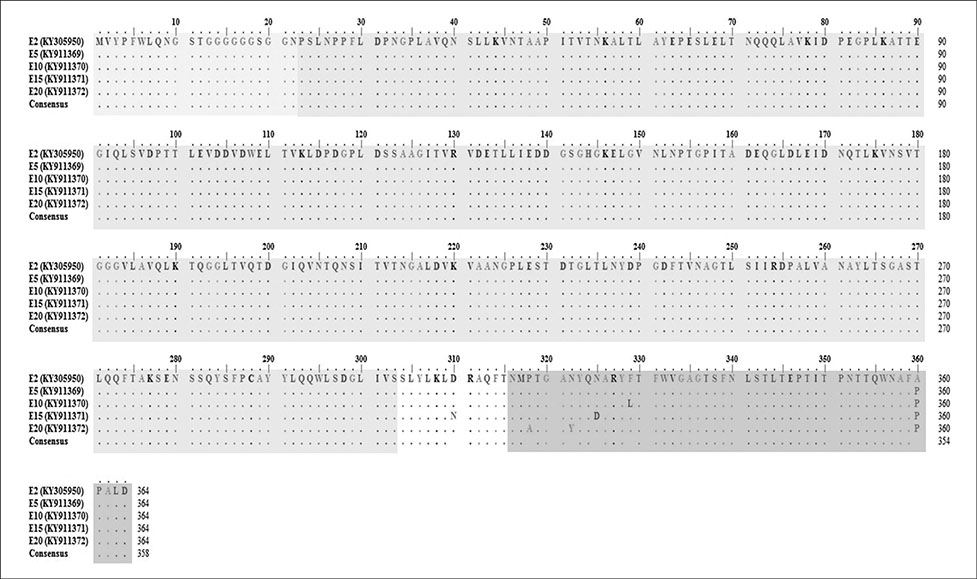
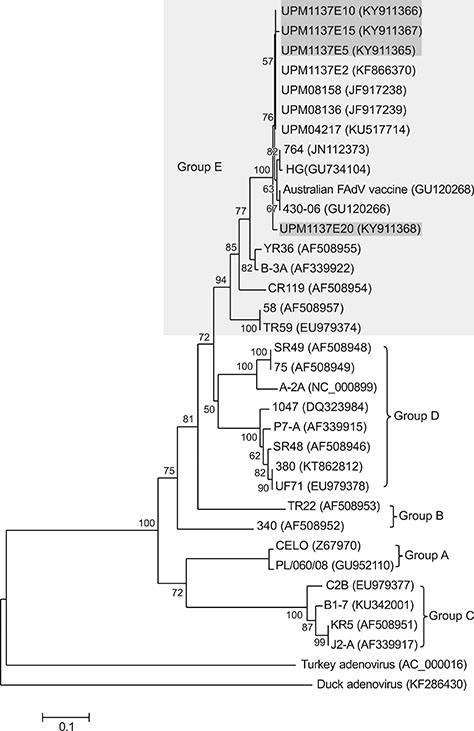
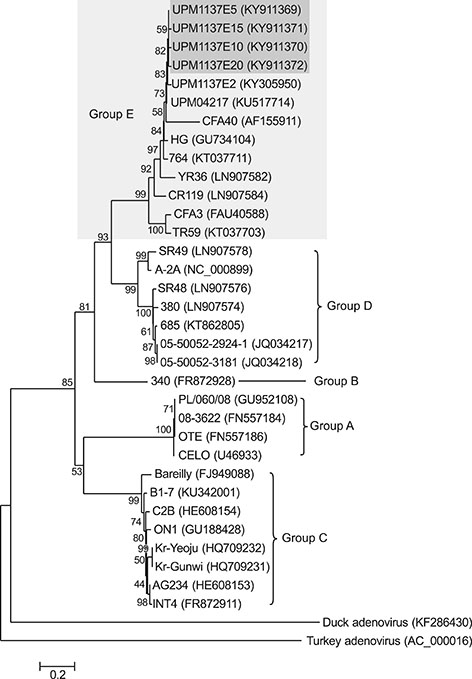
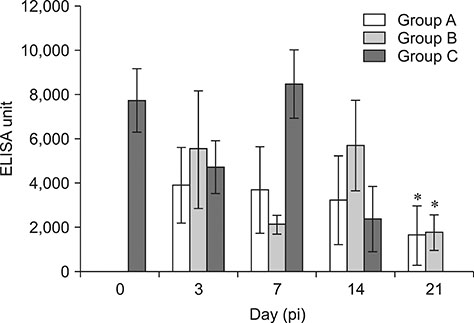
- Fowl adenovirus (FAdV) is distributed worldwide and causes economic losses in the poultry industry. The objectives of this study were to determine the hexon and fiber gene changes in an attenuated FAdV isolate from Malaysia in specific pathogen-free chicken embryonated eggs (SPF CEE) and its infectivity in commercial broiler chickens. SPF CEE were inoculated with 0.1 mL FAdV inoculum via the chorioallantoic membrane (CAM) for 20 consecutive passages. The isolate at passage 20 (E20), with a virus titer of 10(8.7)TCIDâ‚…â‚€/mL (TCIDâ‚…â‚€, 50% tissue culture infective dose), was inoculated (0.5 mL) into one-day-old commercial broiler chicks either via oral or intraperitoneal routes. The study demonstrated that 100% embryonic mortality was recorded from E2 to E20 with a delayed pattern at E17 onwards. The lesions were confined to the liver and CAM. Substitutions of amino acids in the L1 loop of hexon at positions 49 and 66, and in the knob of fiber at positions 318 and 322 were recorded in the E20 isolate. The isolate belongs to serotype 8b and is non-pathogenic to broiler chickens, but it is able to induce a FAdV antibody titer. It appears that molecular changes in the L1 loop of hexon and the knob of fiber are markers for FAdV infectivity.
Keyword
MeSH Terms
Figure
Reference
-
1. Alemnesh W, Hair-Bejo M, Aini I, Omar AR. Pathogenicity of fowl adenovirus in specific pathogen free chicken embryos. J Comp Pathol. 2012; 146:223–229.
Article2. Alvarado IR, Villegas P, El-Attrache J, Jensen E, Rosales G, Perozo F, Purvis LB. Genetic characterization, pathogenicity, and protection studies with an avian adenovirus isolate associated with inclusion body hepatitis. Avian Dis. 2007; 51:27–32.
Article3. Bancroft JD, Stevens A. Theory and Practice of Histological Techniques. 4th ed. Edinburgh: Churchill Livingstone;1996.4. Benko M, Harrach B, Both GW, Russell WC, Adair BM, Adam E, de Jong JC, Hess M, Johnson M. Family Adenoviridae. In : Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses. London: Academic Press;2005. p. 213–228.5. Domanska-Blicharz K, Tomczyk G, Smietanka K, Kozaczynski W, Minta Z. Molecular characterization of fowl adenoviruses isolated from chickens with gizzard erosions. Poult Sci. 2011; 90:983–989.
Article6. Grgić H, Krell PJ, Nagy E. Comparison of fiber gene sequences of inclusion body hepatitis (IBH) and non-IBH strains of serotype 8 and 11 fowl adenoviruses. Virus Genes. 2014; 48:74–80.
Article7. Grgić H, Yang DH, Nagy E. Pathogenicity and complete genome sequence of fowl adenovirus serotype 8 isolate. Virus Res. 2011; 156:91–97.
Article8. Hair-Bejo M. Inclusion body hepatitis in a flock of commercial broiler chickens. J Vet Malaysia. 2005; 17:23–26.9. Itakura C, Matsushita S, Goto M. Fine structure of inclusion bodies in hepatic cells of chickens naturally affected with inclusion body hepatitis. Avian Pathol. 1977; 6:19–32.
Article10. Juliana MA, Nurulfiza I, Hair-Bejo M, Omar AR, Aini I. Molecular characterization of fowl adenoviruses isolated from inclusion body hepatitis outbreaks in commercial broiler chickens in Malaysia. Pertanika J Trop Agric Sci. 2014; 37:483–497.11. Kajan GL, Kecskeméti S, Harrach B, Benkő M. Molecular typing of fowl adenoviruses, isolated in Hungary recently, reveals high diversity. Vet Microbiol. 2013; 167:357–363.
Article12. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007; 23:2947–2948.
Article13. Maartens LH, Joubert HW, Aitchison H, Venter EH. Inclusion body hepatitis associated with an outbreak of fowl adenovirus type 2 and type 8b in broiler flocks in South Africa. J S Afr Vet Assoc. 2014; 85:e1–e5.
Article14. Mansoor MK, Hussain I, Arshad M, Muhammad G. Preparation and evaluation of chicken embryo-adapted fowl adenovirus serotype 4 vaccine in broiler chickens. Trop Anim Health Prod. 2011; 43:331–338.
Article15. Meulemans G, Boschmans M, Berg TP, Decaesstecker M. Polymerase chain reaction combined with restriction enzyme analysis for detection and differentiation of fowl adenoviruses. Avian Pathol. 2001; 30:655–660.
Article16. Morshed R, Hosseini H, Langeroudi AG, Fard MHB, Charkhkar S. Fowl adenoviruses D and E cause inclusion body hepatitis outbreaks in broiler and broiler breeder pullet flocks. Avian Dis. 2017; 61:205–210.
Article17. Norina L, Norsharina A, Nurnadiah AH, Redzuan I, Ardy A, Nor-Ismaliza I. Avian adenovirus isolated from broiler affected with inclusion body hepatitis. Malaysian J Vet Res. 2016; 7:121–126.18. Pallister J, Wright PJ, Sheppard M. A single gene encoding the fiber is responsible for variations in virulence in the fowl adenoviruses. J Virol. 1996; 70:5115–5122.
Article19. Raue R, Hess M. Hexon based PCRs combined with restriction enzyme analysis for rapid detection and differentiation of fowl adenoviruses and egg drop syndrome virus. J Virol Methods. 1998; 73:211–217.
Article20. Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Hyg. 1938; 27:493–497.21. Steer PA, O'Rourke D, Ghorashi SA, Noormohammadi AH. Application of high-resolution melting curve analysis for typing of fowl adenoviruses in field cases of inclusion body hepatitis. Aust Vet J. 2011; 89:184–192.
Article22. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28:2731–2739.
Article23. Wang Z, Wang B, Lou J, Yan J, Gao L, Geng R, Yu B. Mutation in fiber of adenovirus serotype 5 gene therapy vector decreases liver tropism. Int J Clin Exp Med. 2014; 7:4942–4950.24. Zhou H, Zhu J, Tu J, Zou W, Hu Y, Yu Z, Yin W, Li Y, Zhang A, Wu Y, Yu Z, Chen H, Jin M. Effect on virulence and pathogenicity of H5N1 influenza a virus through truncations of NS1 eIF4GI binding domain. J Infect Dis. 2010; 202:1338–1346.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Molecular characterization of Malaysian fowl adenovirus (FAdV) serotype 8b species E and pathogenicity of the virus in specificpathogen-free chicken
- Establishment of a chicken challenge model of fowl adenovirus serotype-4 for vaccine verification
- Isolation and characterization of fowl adenovirus serotype 4 from chickens with hydropericardium syndrome in Korea
- Longevity of Toxocara cati Larvae and Pathology in Tissues of Experimentally Infected Chickens
- Molecular Classification of Human Adenovirus Type 7 Isolated From Acute Respiratory Disease Outbreak (ARD) in Korea, 2005–2006