J Vet Sci.
2018 Sep;19(5):721-724. 10.4142/jvs.2018.19.5.721.
Detection and genetic characterization of porcine circovirus 3 from aborted fetuses and pigs with respiratory disease in Korea
- Affiliations
-
- 1Animal Disease Diagnostic Division, Animal and Plant Quarantine Agency, Gimcheon 39660, Korea.
- 2College of Veterinary Medicine and Animal Disease Intervention Center, Kyungpook National University, Daegu 41566, Korea. parkck@knu.ac.kr
- 3College of Veterinary Medicine, Konkuk University, Seoul 05029, Korea.
- KMID: 2420943
- DOI: http://doi.org/10.4142/jvs.2018.19.5.721
Abstract
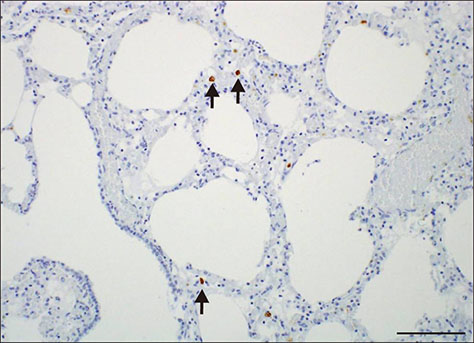
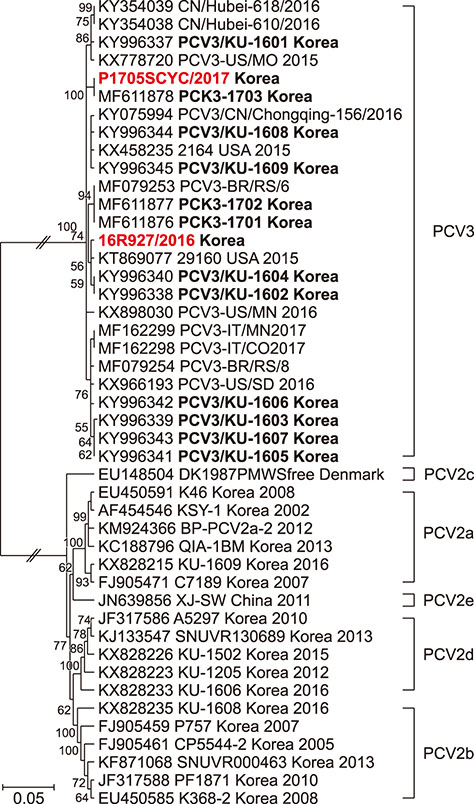
- A novel porcine circovirus 3 (PCV3) was first detected in pigs showing porcine dermatitis and nephropathy syndrome, reproductive failure, and multisystemic inflammation in the USA. Herein, we report on PCV3 as a potential etiological agent of clinical signs, reproductive failure and respiratory distress on Korean pig farms, based on in situ hybridization, pathological, and molecular findings. Confirmation of the presence of PCV3 may increase co-infection with other causative agents of disease in Korean pig herds, indicating the need for further systemic investigation of pathogenicity and of multiple infections with PCV2 genotypes and bacteria, and the development of an effective PCV3 vaccine.
MeSH Terms
Figure
Reference
-
1. Barate AK, Lee HY, Jeong HW, Truong LQ, Joo HG, Hahn TW. An improved multiplex PCR for diagnosis and differentiation of Mycoplasma hyopneumoniae and Mycoplasma hyorhinis. Korean J Vet Res. 2012; 52:39–43.
Article2. Brunborg IM, Jonassen CM, Moldal T, Bratberg B, Lium B, Koenen F, Schonheit J. Association of myocarditis with high viral load of porcine circovirus type 2 in several tissues in cases of fetal death and high mortality in piglets. A case study. J Vet Diagn Invest. 2007; 19:368–375.
Article3. Choi C, Chae C. In-situ hybridization for the detection of porcine circovirus in pigs with postweaning multisystemic wasting syndrome. J Comp Pathol. 1999; 121:265–270.
Article4. Davies B, Wang X, Dvorak CM, Marthaler D, Murtaugh MP. Diagnostic phylogenetics reveals a new Porcine circovirus 2 cluster. Virus Res. 2016; 217:32–37.
Article5. Hamel AL, Lin LL, Nayar GP. Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J Virol. 1998; 72:5262–5267.
Article6. Kang I, Kim D, Han K, Seo HW, Oh Y, Park C, Lee J, Gottschalk M, Chae C. Optimized protocol for multiplex nested polymerase chain reaction to detect and differentiate Haemophilus parasuis, Streptococcus suis, and Mycoplasma hyorhinis in formalin-fixed, paraffin-embedded tissues from pigs with polyserositis. Can J Vet Res. 2012; 76:195–200.7. Kim HH, Park SI, Hyun BH, Park SJ, Jeong YJ, Shin DJ, Chun YH, Hosmillo M, Lee BJ, Kang MI, Cho KO. Genetic diversity of porcine circovirus type 2 in Korean pigs with postweaning multisystemic wasting syndrome during 2005–2007. J Vet Med Sci. 2009; 71:349–353.
Article8. Kwon T, Lee DU, Yoo SJ, Je SH, Shin JY, Lyoo YS. Genotypic diversity of porcine circovirus type 2 (PCV2) and genotype shift to PCV2d in Korean pig population. Virus Res. 2017; 228:24–29.
Article9. Kwon T, Yoo SJ, Park CK, Lyoo YS. Prevalence of novel porcine circovirus 3 in Korean pig populations. Vet Microbiol. 2017; 207:178–180.
Article10. Madson DM, Patterson AR, Ramamoorthy S, Pal N, Meng XJ, Opriessnig T. Reproductive failure experimentally induced in sows via artificial insemination with semen spiked with porcine circovirus type 2. Vet Pathol. 2009; 46:707–716.
Article11. Palinski R, Pineyro P, Shang P, Yuan F, Guo R, Fang Y, Byers E, Hause BM. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. J Virol. 2017; 91:e01879–e01816.
Article12. Phan TG, Giannitti F, Rossow S, Marthaler D, Knutson TP, Li L, Deng X, Resende T, Vannucci F, Delwart E. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol J. 2016; 13:184.
Article13. Segales J. Porcine circovirus type 2 (PCV2) infections: clinical signs, pathology and laboratory diagnosis. Virus Res. 2012; 164:10–19.
Article14. Selim AM, Elhaig MM, Gaede W. Development of multiplex real-time PCR assay for the detection of Brucella spp., Leptospira spp. and Campylobacter foetus. Vet Ital. 2014; 50:269–275.15. Seo HW, Park C, Kang I, Choi K, Jeong J, Park SJ, Chae C. Genetic and antigenic characterization of a newly emerging porcine circovirus type 2b mutant first isolated in cases of vaccine failure in Korea. Arch Virol. 2014; 159:3107–3111.
Article16. Stiles J, Prade R, Greene C. Detection of Toxoplasma gondii in feline and canine biological samples by use of the polymerase chain reaction. Am J Vet Res. 1996; 57:264–267.17. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28:2731–2739.
Article18. Tischer I, Mields W, Wolff D, Vagt M, Griem W. Studies on epidemiology and pathogenicity of porcine circovirus. Arch Virol. 1986; 91:271–276.
Article19. Zhai SL, Chen SN, Wei ZZ, Zhang JW, Huang L, Lin T, Yue C, Ran DL, Yuan SS, Wei WK, Long JX. Co-existence of multiple strains of porcine circovirus type 2 in the same pig from China. Virol J. 2011; 8:517.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Prevalence of porcine reproductive and respiratory syndrome virus, porcine circovirus type 2 and porcine parvovirus from aborted fetuses and pigs with respiratory problems in Korea
- First detection and genetic characterization of porcine parvovirus 7 from Korean domestic pig farms
- Pathologic studies in lymph nodes of pigs infected with porcine circovirus type 2, porcine reproductive and respiratory syndrome virus
- Prevalence and pathologic study of porcine salmonellosis in Jeju
- Pathological characteristics on porcine enteritis associated with porcine circovirus type 2 in Jeju