J Korean Med Sci.
2017 Jan;32(1):38-46. 10.3346/jkms.2017.32.1.38.
BRAF-Mutated Colorectal Cancer Exhibits Distinct Clinicopathological Features from Wild-Type BRAF-Expressing Cancer Independent of the Microsatellite Instability Status
- Affiliations
-
- 1Department of Pathology, Konkuk University School of Medicine, Seoul, Korea. 20040002@kuh.ac.kr
- 2Department of Surgery, Konkuk University School of Medicine, Seoul, Korea.
- KMID: 2417583
- DOI: http://doi.org/10.3346/jkms.2017.32.1.38
Abstract
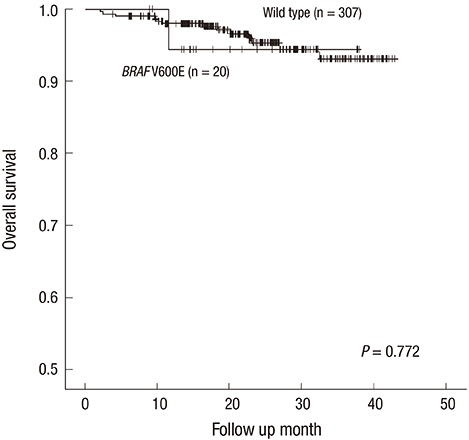
- In patients with colorectal cancer (CRC), the BRAF V600E mutation has been reported to be associated with several clinicopathological features and poor survival. However, the prognostic implications of BRAF V600E mutation and the associated clinicopathological characteristics in CRCs remain controversial. Therefore, we reviewed various clinicopathological features, including BRAF status, in 349 primary CRCs and analyzed the relationship between BRAF status and various clinicopathological factors, including overall survival. Similar to previous studies conducted in Eastern countries, the incidence of the BRAF V600E mutation in the current study was relatively low (5.7%). BRAF-mutated CRC exhibits distinct clinicopathological features from wild-type BRAF-expressing cancer independent of the microsatellite instability (MSI) status. This mutation was significantly associated with a proximal tumor location (P = 0.002); mucinous, signet ring cell, and serrated tumor components (P < 0.001, P = 0.003, and P = 0.008, respectively); lymphovascular invasion (P = 0.004); a peritumoral lymphoid reaction (P = 0.009); tumor budding (P = 0.046); and peritoneal seeding (P = 0.012). In conclusion, the incidence of the BRAF V600E mutation was relatively low in this study. BRAF-mutated CRCs exhibited some clinicopathological features which were also frequently observed in MSI-H CRCs, such as a proximal location; mucinous, signet ring cell, and serrated components; and marked peritumoral lymphoid reactions.
MeSH Terms
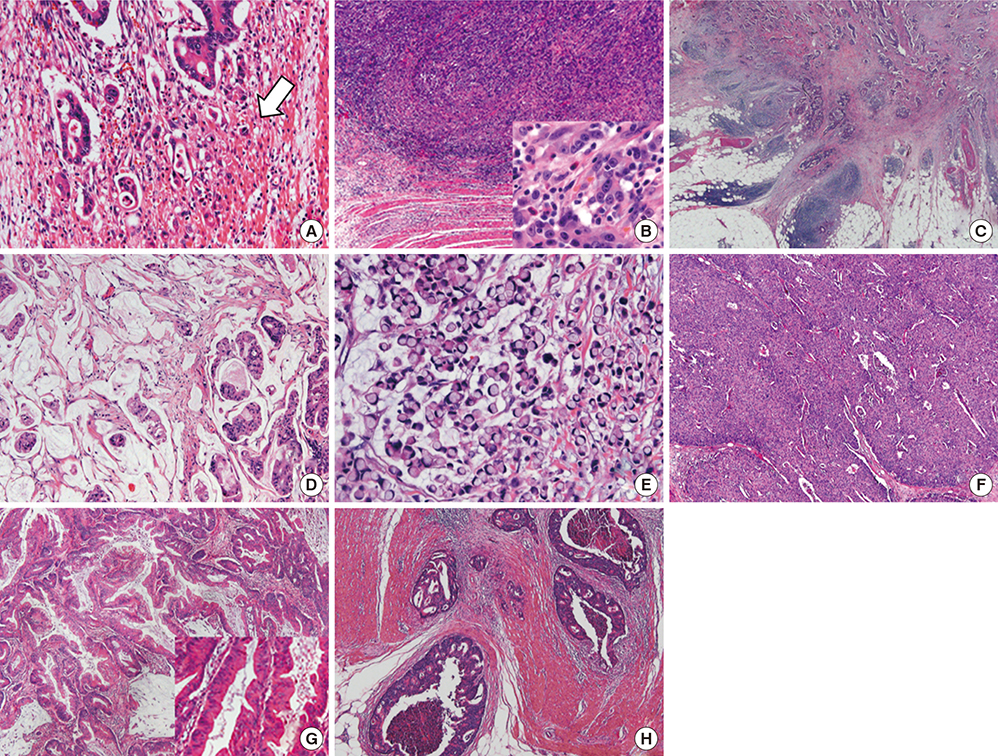
Figure
Reference
-
1. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005; 55:74–108.2. National Cancer Information Center (KR). Cancer Incidence trend analysis [Internet]. accessed on 7 June 2015. Available at http://www.cancer.go.kr/mbs/cancer/subview.jsp?id=cancer_040201000000.3. Ikenoue T, Hikiba Y, Kanai F, Tanaka Y, Imamura J, Imamura T, Ohta M, Ijichi H, Tateishi K, Kawakami T, et al. Functional analysis of mutations within the kinase activation segment of B-Raf in human colorectal tumors. Cancer Res. 2003; 63:8132–8137.4. Zlobec I, Bihl MP, Schwarb H, Terracciano L, Lugli A. Clinicopathological and protein characterization of BRAF- and K-RAS-mutated colorectal cancer and implications for prognosis. Int J Cancer. 2010; 127:367–380.5. Bogaert J, Prenen H. Molecular genetics of colorectal cancer. Ann Gastroenterol. 2014; 27:9–14.6. Mao C, Liao RY, Chen Q. BRAF mutation predicts resistance to anti-EGFR monoclonal antibodies in wild-type KRAS metastatic colorectal cancer. J Cancer Res Clin Oncol. 2010; 136:1293–1294.7. Yuan ZX, Wang XY, Qin QY, Chen DF, Zhong QH, Wang L, Wang JP. The prognostic role of BRAF mutation in metastatic colorectal cancer receiving anti-EGFR monoclonal antibodies: a meta-analysis. PLoS One. 2013; 8:e65995.8. Cui D, Cao D, Yang Y, Qiu M, Huang Y, Yi C. Effect of BRAF V600E mutation on tumor response of anti-EGFR monoclonal antibodies for first-line metastatic colorectal cancer treatment: a meta-analysis of randomized studies. Mol Biol Rep. 2014; 41:1291–1298.9. Herr R, Köhler M, Andrlová H, Weinberg F, Möller Y, Halbach S, Lutz L, Mastroianni J, Klose M, Bittermann N, et al. B-Raf inhibitors induce epithelial differentiation in BRAF-mutant colorectal cancer cells. Cancer Res. 2015; 75:216–229.10. Yaeger R, Saltz LB. RAF plus EGFR inhibition for BRAF-mutant metastatic colorectal cancer-response. Clin Cancer Res. 2015; 21:2188.11. Thiel A, Ristimäki A. Toward a molecular classification of colorectal cancer: the role of BRAF. Front Oncol. 2013; 3:281.12. French AJ, Sargent DJ, Burgart LJ, Foster NR, Kabat BF, Goldberg R, Shepherd L, Windschitl HE, Thibodeau SN. Prognostic significance of defective mismatch repair and BRAF V600E in patients with colon cancer. Clin Cancer Res. 2008; 14:3408–3415.13. Mouradov D, Domingo E, Gibbs P, Jorissen RN, Li S, Soo PY, Lipton L, Desai J, Danielsen HE, Oukrif D, et al. Survival in stage II/III colorectal cancer is independently predicted by chromosomal and microsatellite instability, but not by specific driver mutations. Am J Gastroenterol. 2013; 108:1785–1793.14. Kim JH, Bae JM, Oh HJ, Lee HS, Kang GH. Pathologic factors associated with prognosis after adjuvant chemotherapy in stage II/III microsatellite-unstable colorectal cancers. J Pathol Transl Med. 2015; 49:118–128.15. Ueno H, Mochizuki H, Hashiguchi Y, Shimazaki H, Aida S, Hase K, Matsukuma S, Kanai T, Kurihara H, Ozawa K, et al. Risk factors for an adverse outcome in early invasive colorectal carcinoma. Gastroenterology. 2004; 127:385–394.16. Klintrup K, Mäkinen JM, Kauppila S, Väre PO, Melkko J, Tuominen H, Tuppurainen K, Mäkelä J, Karttunen TJ, Mäkinen MJ. Inflammation and prognosis in colorectal cancer. Eur J Cancer. 2005; 41:2645–2654.17. Kaserer K, Schmaus J, Bethge U, Migschitz B, Fasching S, Walch A, Herbst F, Teleky B, Wrba F. Staining patterns of p53 immunohistochemistry and their biological significance in colorectal cancer. J Pathol. 2000; 190:450–456.18. Chung HW, Lee SY, Han HS, Park HS, Yang JH, Lee HH, So Y. Gastric cancers with microsatellite instability exhibit high fluorodeoxyglucose uptake on positron emission tomography. Gastric Cancer. 2013; 16:185–192.19. Kim SK, Kim DL, Han HS, Kim WS, Kim SJ, Moon WJ, Oh SY, Hwang TS. Pyrosequencing analysis for detection of a BRAFV600E mutation in an FNAB specimen of thyroid nodules. Diagn Mol Pathol. 2008; 17:118–125.20. Kim JH, Kang GH. Molecular and prognostic heterogeneity of microsatellite-unstable colorectal cancer. World J Gastroenterol. 2014; 20:4230–4243.21. Kang GH. Four molecular subtypes of colorectal cancer and their precursor lesions. Arch Pathol Lab Med. 2011; 135:698–703.22. Chen D, Huang JF, Liu K, Zhang LQ, Yang Z, Chuai ZR, Wang YX, Shi DC, Huang Q, Fu WL. BRAFV600E mutation and its association with clinicopathological features of colorectal cancer: a systematic review and meta-analysis. PLoS One. 2014; 9:e90607.23. Sinicrope FA, Rego RL, Foster N, Sargent DJ, Windschitl HE, Burgart LJ, Witzig TE, Thibodeau SN. Microsatellite instability accounts for tumor site-related differences in clinicopathologic variables and prognosis in human colon cancers. Am J Gastroenterol. 2006; 101:2818–2825.24. Lavotshkin S, Jalas JR, Torisu-Itakura H, Ozao-Choy J, Lee JH, Sim MS, Stojadinovic A, Wainberg Z, Bifulco CB, Fox BA, et al. Immunoprofiling for prognostic assessment of colon cancer: a novel complement to ultrastaging. J Gastrointest Surg. 2015; 19:999–1006.25. Gatalica Z, Torlakovic E. Pathology of the hereditary colorectal carcinoma. Fam Cancer. 2008; 7:15–26.26. Jenkins MA, Hayashi S, O’Shea AM, Burgart LJ, Smyrk TC, Shimizu D, Waring PM, Ruszkiewicz AR, Pollett AF, Redston M, et al. Pathology features in Bethesda guidelines predict colorectal cancer microsatellite instability: a population-based study. Gastroenterology. 2007; 133:48–56.27. Yoon HH, Orrock JM, Foster NR, Sargent DJ, Smyrk TC, Sinicrope FA. Prognostic impact of FoxP3+ regulatory T cells in relation to CD8+ T lymphocyte density in human colon carcinomas. PLoS One. 2012; 7:e42274.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Differential Sensitivity of Wild-Type and BRAF-Mutated Cells to Combined BRAF and Autophagy Inhibition
- Immune check point inhibitors in BRAF mutated metastatic colorectal cancer: A review
- Utility of BRAF VE1 Immunohistochemistry as a Screening Tool for Colorectal Cancer Harboring BRAF V600E Mutation
- hMLH1 promoter methylation and BRAF mutations in high-frequency microsatellite instability colorectal cancers not fulfilling the revised Bethesda guidelines
- Hyperplastic Polyposis Syndrome Identified with a BRAF Mutation