J Vet Sci.
2016 Dec;17(4):523-529. 10.4142/jvs.2016.17.4.523.
Molecular and phylogenetic analysis of Anaplasma spp. in sheep and goats from six provinces of China
- Affiliations
-
- 1College of Animal Science and Veterinary Medicine, Henan Agricultural University, Zhengzhou 450002, China. zhengqingchun1234@126.com
- KMID: 2412608
- DOI: http://doi.org/10.4142/jvs.2016.17.4.523
Abstract
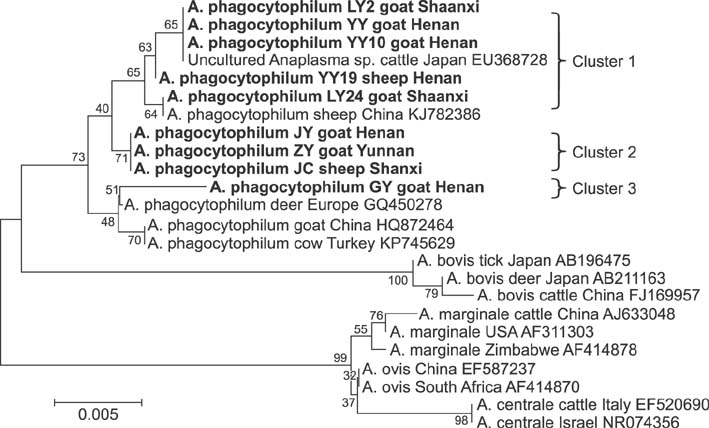
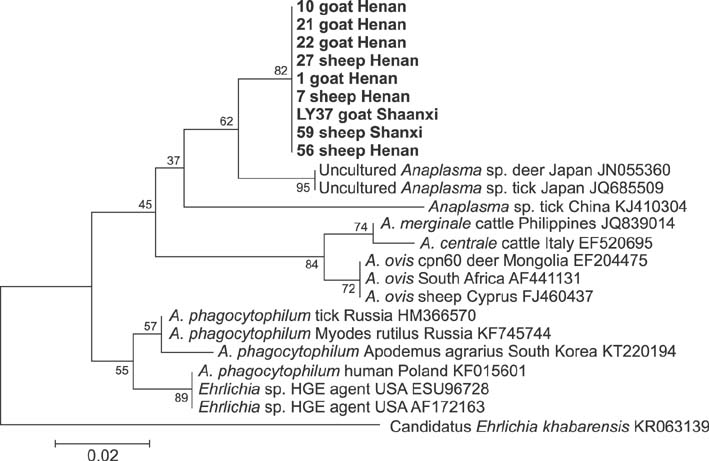
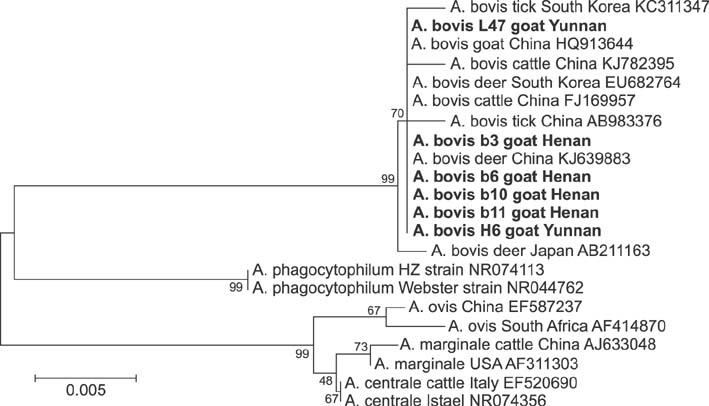
- Members of the genus Anaplasma are important emerging tick-borne pathogens in both humans and animals in tropical and subtropical areas. Here, we investigated the presence of Anaplasma spp. in 621 sheep and 710 goats from six provinces of China. Polymerase chain reaction (PCR) and DNA sequencing were conducted to determine the prevalence of Anaplasma (A.) phagocytophilum, A. ovis and A. bovis targeting the 16S ribosomal RNA or the major surface protein 4 gene. PCR revealed Anaplasma in 39.0% (240/621) of sheep and 45.5% (323/710) of goats. The most frequently detected species was A. ovis (88/621, 14.2% for sheep; 129/710, 18.2% for goats), followed by A. bovis (60/621, 9.7% for sheep; 74/710, 10.4% for goats) and A. phagocytophilum (33/621, 5.3% for sheep; 15/710, 2.1% for goats). Additionally, eight sheep and 20 goats were found to be infected with three pathogens simultaneously. DNA sequencing confirmed the presence of these three Anaplasma species in the investigated areas, and phylogenetic analysis indicated that there was geographic segregation to a certain extent, as well as a relationship between the host and cluster of A. ovis. The results of the present study provide valuable data that helps understand the epidemiology of anaplasmosis in ruminants from China.
Keyword
MeSH Terms
-
Anaplasma/genetics/*physiology
Anaplasmosis/*epidemiology/parasitology
Animals
Chaperonin 60/genetics
China/epidemiology
DNA, Protozoan/genetics
Female
Goat Diseases/*epidemiology
Goats
Phylogeny
Polymerase Chain Reaction/veterinary
Protozoan Proteins/genetics
RNA, Ribosomal, 16S/genetics
Sequence Analysis, DNA
Sheep
Sheep Diseases/*epidemiology/parasitology
Species Specificity
Chaperonin 60
DNA, Protozoan
Protozoan Proteins
RNA, Ribosomal, 16S
Figure
Reference
-
1. Barlough JE, Madigan JE, DeRock E, Bigornia L. Nested polymerase chain reaction for detection of Ehrlichia equi genomic DNA in horses and ticks (Ixodes pacificus). Vet Parasitol. 1996; 63:319–329.
Article2. de la Fuente J, Atkinson MW, Naranjo V, Fernández de Mera IG, Mangold AJ, Keating KA, Kocan KM. Sequence analysis of the msp4 gene of Anaplasma ovis strains. Vet Microbiol. 2007; 119:375–381.3. Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, Rikihisa Y, Rurangirwa FR. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int J Syst Evol Microbiol Int J Syst Evol Microbiol. 2001; 51:2145–2165.4. Hassan MA, Raoofi A, Lotfollahzadeh S, Javanbakht J. Clinical and cytological characteristics and prognostic implications on sheep and goat Theileria infection in north of Iran. J Parasit Dis. 2015; 39:190–193.
Article5. Kawahara M, Rikihisa Y, Lin Q, Isogai E, Tahara K, Itagaki A, Hiramitsu Y, Tajima T. Novel genetic variants of Anaplasma phagocytophilum, Anaplasma bovis, Anaplasma centrale, and a novel Ehrlichia sp. in wild deer and ticks on two major islands in Japan. Appl Environ Microbiol. 2006; 72:1102–1109.
Article6. Kumar S, Tamura K, Nei M. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput Appl Biosci. 1994; 10:189–191.
Article7. Liu Z, Ma M, Wang Z, Wang J, Peng Y, Li Y, Guan G, Luo J, Yin H. Molecular survey and genetic identification of Anaplasma species in goats from central and southern China. Appl Environ Microbiol. 2012; 78:464–470.
Article8. Lommano E, Dvořák C, Vallotton L, Jenni L, Gern L. Tick-borne pathogens in ticks collected from breeding and migratory birds in Switzerland. Ticks Tick Borne Dis. 2014; 5:871–882.
Article9. Ooshiro M, Zakimi S, Matsukawa Y, Katagiri Y, Inokuma H. Detection of Anaplasma bovis and Anaplasma phagocytophilum from cattle on Yonaguni Island, Okinawa, Japan. Vet Parasitol. 2008; 154:360–364.
Article10. Rar V, Golovljova I. Anaplasma, Ehrlichia, and "Candidatus Neoehrlichia" bacteria: pathogenicity, biodiversity, and molecular genetic characteristics, a review. Infect Genet Evol. 2011; 11:1842–1861.
Article11. Rymaszewska A. Genotyping of Anaplasma phagocytophilum strains from Poland for selected genes. Folia Biol (Krakow). 2014; 62:37–48.12. Sainz A, Amusategui I, Tesouro MA. Ehrlichia platys infection and disease in dogs in Spain. J Vet Diagn Invest. 1999; 11:382–384.
Article13. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987; 4:406–425.14. Şevik M, Sait A. Genetic characterization of peste des petits ruminants virus, Turkey, 2009-2013. Res Vet Sci. 2015; 101:187–195.
Article15. Singh G, Singh R, Verma PK, Anand A. Anthelmintic efficacy of aqueous extract of Butea monosperma (Lam.) Kuntze against Haemonchus contortus of sheep and goats. J Parasit Dis. 2015; 39:200–205.
Article16. Sreekumar C, Anandan R, Balasundaram S, Rajavelu G. Morphology and staining characteristics of Ehrlichia bovis. Comp Immunol Microbiol Infect Dis. 1996; 19:79–83.17. Stuen S, Granquist EG, Silaghi C. Anaplasma phagocytophilum—a widespread multi-host pathogen with highly adaptive strategies. Front Cell Infect Microbiol. 2013; 3:31.18. Stuen S, Whist SK, Bergström K, Moum T. Possible exclusion of genotypes in Anaplasma phagocytophilum-infected lambs. Vet Rec. 2005; 156:518–520.19. Wareth G, Melzer F, Tomaso H, Roesler U, Neubauer H. Detection of Brucella abortus DNA in aborted goats and sheep in Egypt by real-time PCR. BMC Res Notes. 2015; 8:212.20. Yang J, Li Y, Liu Z, Liu J, Niu Q, Ren Q, Chen Z, Guan G, Luo J, Yin H. Molecular detection and characterization of Anaplasma spp. in sheep and cattle from Xinjiang, northwest China. Parasit Vectors. 2015; 8:108.21. Zhan L, Cao WC, Jiang JF, Zhang XA, Liu YX, Wu XM, Zhang WY, Zhang PH, Bian CL, Dumler JS, Yang H, Zuo SQ, Chu CY, Liu W, Richardus JH, Habbema JD. Anaplasma phagocytophilum from Rodents and Sheep, China. Emerg Infect Dis. 2010; 16:764–768.22. Zhan L, Cao WC, Jiang JF, Zhang XA, Wu XM, Zhang WY, Liu W, Zuo SQ, Cao ZW, Yang H, Richardus JH, Habbema JD. Anaplasma phagocytophilum in livestock and small rodents. Vet Microbiol. 2010; 144:405–408.23. Zhan L, Chu CY, Zuo SQ, Wu XM, Dumler JS, Jia N, Jiang BG, Yang H, Cao WC. Anaplasma phagocytophilum and Borrelia burgdorferi in rabbits from southeastern China. Vet Parasitol. 2009; 162:354–356.
Article24. Zhang Y, Lv Y, Cui Y, Wang J, Cao S, Jian F, Wang R, Zhang L, Ning C. First molecular evidence for the presence of Anaplasma DNA in milk from sheep and goats in China. Parasitol Res. 2016; 115:2789–2795.
Article25. Zhou Z, Nie K, Tang C, Wang Z, Zhou R, Hu S, Zhang Z. Phylogenetic analysis of the genus Anaplasma in southwestern China based on 16S rRNA sequence. Res Vet Sci. 2010; 89:262–265.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Detection of Anaplasma sp. in Korean Native Goats (Capra aegagrus hircus) on Jeju Island, Korea
- Serological Detection of Antibodies against Anaplasma spp. in Cattle Reared in the Gyeongsangbuk-do, Korea
- Phylogenetic Analysis of Ruminant Theileria spp. from China Based on 28S Ribosomal RNA Gene
- Identification and prevalence of Ehrlichia chaffeensis infection in Haemaphysalis longicornis ticks from Korea by PCR, sequencing and phylogenetic analysis based on 16S rRNA gene
- Prevalence of intestinal parasites in animal hosts and potential implications to animal and human health in Edo, Nigeria