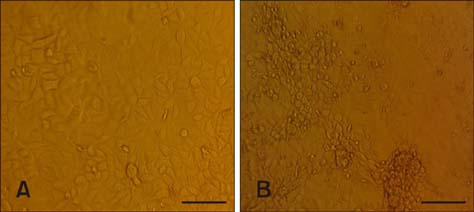
J Vet Sci.
2018 May;19(3):339-349. 10.4142/jvs.2018.19.3.339.
Genomic characterization and pathogenic study of two porcine reproductive and respiratory syndrome viruses with different virulence in Fujian, China
- Affiliations
-
- 1Institute of Animal Sciences, Chinese Academy of Agricultural Sciences, Beijing 100193, China. houshaohua@126.com
- 2Molecular and Cellular Biology, Gembloux Agro-Bio Tech University of Liège, 5030 Gembloux, Belgium.
- KMID: 2412124
- DOI: http://doi.org/10.4142/jvs.2018.19.3.339
Abstract
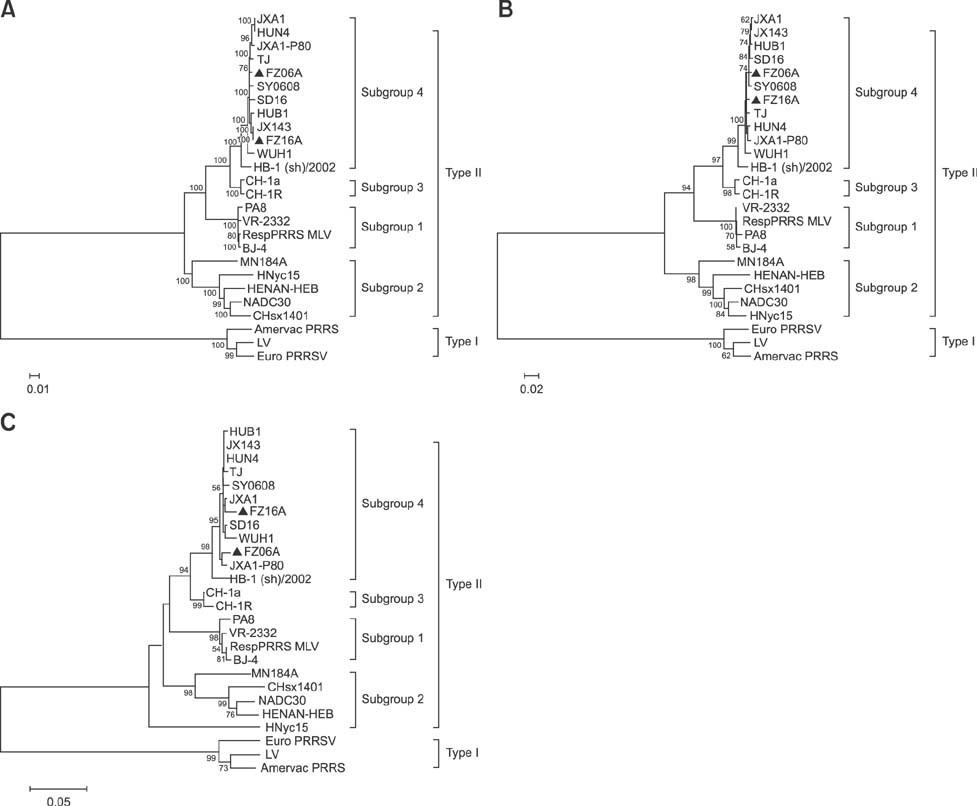
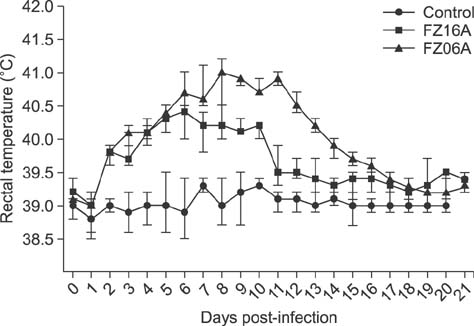
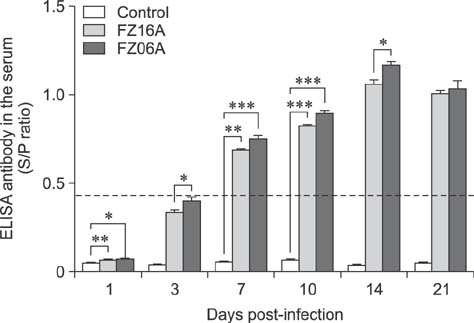
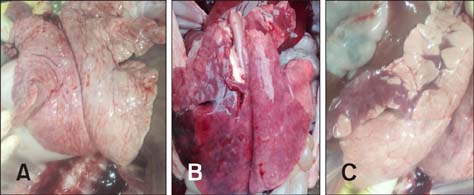
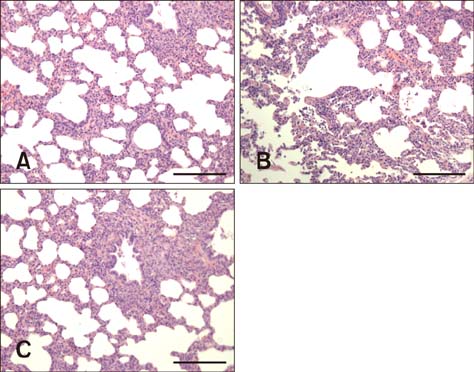
- Two strains of porcine reproductive and respiratory syndrome virus (PRRSV) were isolated in 2006 and 2016 and designated as FZ06A and FZ16A, respectively. Inoculation experiments showed that FZ06A caused 100% morbidity and 60% mortality, while FZ16A caused 100% morbidity without death. By using genomic sequence and phylogenetic analyses, close relationships between a Chinese highly pathogenic PRRSV strain and the FZ06A and FZ16A strains were observed. Based on the achieved results, multiple genomic variations in Nsp2, a unique N-glycosylation site (N³³â†’K³³), and a K151 amino acid (AA) substitution for virulence in the GP5 of FZ16A were detected; except the 30 AA deletion in the Nsp2-coding region. Inoculation experiments were conducted and weaker virulence of FZ16A than FZ06A was observed. Based on our results, a 30 AA deletion in the Nsp2-coding region is an unreliable genomic indicator of a high virulence PRRSV strain. The Nsp2 and GP5 differences, in addition to the virulence difference between these two highly pathogenic PRRSV strains, have the potential to be used to establish a basis for further study of PRRSV virulence determinants and to provide data useful in the development of vaccines against this economically devastating disease.
MeSH Terms
Figure
Reference
-
1. Allende R, Kutish GF, Laegreid W, Lu Z, Lewis TL, Rock DL, Friesen J, Galeota JA, Doster AR, Osorio FA. Mutations in the genome of porcine reproductive and respiratory syndrome virus responsible for the attenuation phenotype. Arch Virol. 2000; 145:1149–1161.
Article2. An TQ, Tian ZJ, Leng CL, Peng JM, Tong GZ. Highly pathogenic porcine reproductive and respiratory syndrome virus, Asia. Emerg Infect Dis. 2011; 17:1782–1784.
Article3. Ansari IH, Kwon B, Osorio FA, Pattnaik AK. Influence of N-linked glycosylation of porcine reproductive and respiratory syndrome virus GP5 on virus infectivity, antigenicity, and ability to induce neutralizing antibodies. J Virol. 2006; 80:3994–4004.
Article4. Benfield DA, Nelson E, Collins JE, Harris L, Goyal SM, Robison D, Christianson WT, Morrison RB, Gorcyca D, Chladek D. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332). J Vet Diagn Invest. 1992; 4:127–133.
Article5. Chen Z, Zhou X, Lunney JK, Lawson S, Sun Z, Brown E, Christopher-Hennings J, Knudsen D, Nelson E, Fang Y. Immunodominant epitopes in nsp2 of porcine reproductive and respiratory syndrome virus are dispensable for replication, but play an important role in modulation of the host immune response. J Gen Virol. 2010; 91:1047–1057.
Article6. Collins JE, Benfield DA, Christianson WT, Harris L, Hennings JC, Shaw DP, Goyal SM, McCullough S, Morrison RB, Joo HS, Gorcyca D, Chladek D. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J Vet Diagn Invest. 1992; 4:117–126.
Article7. Dea S, Gagnon CA, Mardassi H, Pirzadeh B, Rogan D. Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the North American and European isolates. Arch Virol. 2000; 145:659–688.
Article8. Du Y, Lu Y, Qi J, Wu J, Wang G, Wang J. Complete genome sequence of a moderately pathogenic porcine reproductive and respiratory syndrome virus variant strain. J Virol. 2012; 86:13883–13884.
Article9. Fan B, Wang H, Bai J, Zhang L, Jiang P. A novel isolate with deletion in GP3 gene of porcine reproductive and respiratory syndrome virus from mid-eastern China. Biomed Res Int. 2014; 2014:306130.10. Fang Y, Fang L, Wang Y, Lei Y, Luo R, Wang D, Chen H, Xiao S. Porcine reproductive and respiratory syndrome virus nonstructural protein 2 contributes to NF-κB activation. Virol J. 2012; 9:83.
Article11. Fang Y, Kim DY, Ropp S, Steen P, Christopher-Hennings J, Nelson EA, Rowland RR. Heterogeneity in Nsp2 of European-like porcine reproductive and respiratory syndrome viruses isolated in the United States. Virus Res. 2004; 100:229–235.
Article12. Firth AE, Zevenhoven-Dobbe JC, Wills NM, Go YY, Balasuriya UB, Atkins JF, Snijder EJ, Posthuma CC. Discovery of a small arterivirus gene that overlaps the GP5 coding sequence and is important for virus production. J Gen Virol. 2011; 92:1097–1106.
Article13. Forsberg R, Storgaard T, Nielsen HS, Oleksiewicz MB, Cordioli P, Sala G, Hein J, Bøtner A. The genetic diversity of European type PRRSV is similar to that of the North American type but is geographically skewed within Europe. Virology. 2002; 299:38–47.
Article14. Halbur PG, Paul PS, Frey ML, Landgraf J, Eernisse K, Meng XJ, Lum MA, Andrews JJ, Rathje JA. Comparison of the pathogenicity of two US porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet Pathol. 1995; 32:648–660.
Article15. Huang C, Zhang Q, Feng WH. Regulation and evasion of antiviral immune responses by porcine reproductive and respiratory syndrome virus. Virus Res. 2015; 202:101–111.
Article16. Jiang P, Chen PY, Dong YY, Cai JL, Cai BX, Jiang ZH. Isolation and genome characterization of porcine reproductive and respiratory syndrome virus in P.R. China. J Vet Diagn Invest. 2000; 12:156–158.
Article17. Johnson CR, Griggs TF, Gnanandarajah J, Murtaugh MP. Novel structural protein in porcine reproductive and respiratory syndrome virus encoded by an alternative ORF5 present in all arteriviruses. J Gen Virol. 2011; 92:1107–1116.
Article18. Karniychuk UU, Geldhof M, Vanhee M, Van Doorsselaere J, Saveleva TA, Nauwynck HJ. Pathogenesis and antigenic characterization of a new East European subtype 3 porcine reproductive and respiratory syndrome virus isolate. BMC Vet Res. 2010; 6:30.
Article19. Ladinig A, Detmer SE, Clarke K, Ashley C, Rowland RR, Lunney JK, Harding JC. Pathogenicity of three type 2 porcine reproductive and respiratory syndrome virus strains in experimentally inoculated pregnant gilts. Virus Res. 2015; 203:24–35.
Article20. Li B, Xiao S, Wang Y, Xu S, Jiang Y, Chen H, Fang L. Immunogenicity of the highly pathogenic porcine reproductive and respiratory syndrome virus GP5 protein encoded by a synthetic ORF5 gene. Vaccine. 2009; 27:1957–1963.
Article21. Li Y, Xue C, Wang L, Chen X, Chen F, Cao Y. Genomic analysis of two Chinese strains of porcine reproductive and respiratory syndrome viruses with different virulence. Virus Genes. 2010; 40:374–381.
Article22. Ma H, Kien F, Manière M, Zhang Y, Lagarde N, Tse KS, Poon LL, Nal B. Human annexin A6 interacts with influenza a virus protein M2 and negatively modulates infection. J Virol. 2012; 86:1789–1801.
Article23. Meulenberg JJ. PRRSV, the virus. Vet Res. 2000; 31:11–21.
Article24. Murtaugh MP, Faaberg KS, Laber J, Elam M, Kapur V. Genetic variation in the PRRS virus. Adv Exp Med Biol. 1998; 440:787–794.
Article25. Nelsen CJ, Murtaugh MP, Faaberg KS. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J Virol. 1999; 73:270–280.
Article26. Ni J, Yang S, Bounlom D, Yu X, Zhou Z, Song J, Khamphouth V, Vatthana T, Tian K. Emergence and pathogenicity of highly pathogenic Porcine reproductive and respiratory syndrome virus in Vientiane, Lao People's Democratic Republic. J Vet Diagn Invest. 2012; 24:349–354.
Article27. Ostrowski M, Galeota JA, Jar AM, Platt KB, Osorio FA, Lopez OJ. Identification of neutralizing and nonneutralizing epitopes in the porcine reproductive and respiratory syndrome virus GP5 ectodomain. J Virol. 2002; 76:4241–4250.
Article28. Paton DJ, Brown IH, Edwards S, Wensvoort G. ‘Blue ear’ disease of pigs. Vet Rec. 1991; 128:617.
Article29. Shi M, Lam TT, Hon CC, Murtaugh MP, Davies PR, Hui RK, Li J, Wong LT, Yip CW, Jiang JW, Leung FC. Phylogeny-based evolutionary, demographical, and geographical dissection of North American type 2 porcine reproductive and respiratory syndrome viruses. J Virol. 2010; 84:8700–8711.
Article30. Snijder EJ, Meulenberg JJ. The molecular biology of arteriviruses. J Gen Virol. 1998; 79:961–979.
Article31. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28:2731–2739.
Article32. Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C, Hu Y, Chen X, Hu D, Tian X, Liu D, Zhang S, Deng X, Ding Y, Yang L, Zhang Y, Xiao H, Qiao M, Wang B, Hou L, Wang X, Yang X, Kang L, Sun M, Jin P, Wang S, Kitamura Y, Yan J, Gao GF. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One. 2007; 2:e526.
Article33. Tong GZ, Zhou YJ, Hao XF, Tian ZJ, An TQ, Qiu HJ. Highly pathogenic porcine reproductive and respiratory syndrome, China. Emerg Infect Dis. 2007; 13:1434–1436.
Article34. Vu HL, Kwon B, Yoon KJ, Laegreid WW, Pattnaik AK, Osorio FA. Immune evasion of porcine reproductive and respiratory syndrome virus through glycan shielding involves both glycoprotein 5 as well as glycoprotein 3. J Virol. 2011; 85:5555–5564.
Article35. Wang FX, Song N, Chen LZ, Cheng SP, Wu H, Wen YJ. Non-structural protein 2 of the porcine reproductive and respiratory syndrome (PRRS) virus: a crucial protein in viral pathogenesis, immunity and diagnosis. Res Vet Sci. 2013; 95:1–7.
Article36. Wang X, Marthaler D, Rovira A, Rossow S, Murtaugh MP. Emergence of a virulent porcine reproductive and respiratory syndrome virus in vaccinated herds in the United States. Virus Res. 2015; 210:34–41.
Article37. Zhang G, Lu W, Chen Y, Zhu L, Wei Z, Li Z, Sun B, Xie Q, Bi Y, Ma J. Complete genome sequence of two variant porcine reproductive and respiratory syndrome viruses isolated from vaccinated piglets. J Virol. 2012; 86:11396–11397.
Article38. Zhou L, Chen S, Zhang J, Zeng J, Guo X, Ge X, Zhang D, Yang H. Molecular variation analysis of porcine reproductive and respiratory syndrome virus in China. Virus Res. 2009; 145:97–105.
Article39. Zhou L, Zhang J, Zeng J, Yin S, Li Y, Zheng L, Guo X, Ge X, Yang H. The 30-amino-acid deletion in the Nsp2 of highly pathogenic porcine reproductive and respiratory syndrome virus emerging in China is not related to its virulence. J Virol. 2009; 83:5156–5167.
Article40. Zhou Z, Li X, Liu Q, Hu D, Yue X, Ni J, Yu X, Zhai X, Galliher-Beckley A, Chen N, Shi J, Tian K. Complete genome sequence of two novel Chinese virulent porcine reproductive and respiratory syndrome virus variants. J Virol. 2012; 86:6373–6374.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Intracellular Localization of the Porcine Reproductive and Respiratory Syndrome Virus Nucleocapsid Protein
- Genetic diversity and phylogenetic analysis of porcine reproductive and respiratory syndrome virus in southern China from 2007 to 2014
- Prevalence of porcine reproductive and respiratory syndrome virus, porcine circovirus type 2 and porcine parvovirus from aborted fetuses and pigs with respiratory problems in Korea
- A new recombined porcine reproductive and respiratory syndrome virus virulent strain in China
- 5' and 3' cis-Acting RNA Elements Required for RNA Replication of Porcine Reproductive and Respiratory Syndrome Virus