Lab Anim Res.
2017 Jun;33(2):49-56. 10.5625/lar.2017.33.2.49.
Quinolone susceptibility and genetic characterization of Salmonella enterica subsp. enterica isolated from pet turtles
- Affiliations
-
- 1Veterinary Medical Center and College of Veterinary Medicine, Chungbuk National University, Cheongju 28644, Korea. gjheo@cbu.ac.kr
- KMID: 2407416
- DOI: http://doi.org/10.5625/lar.2017.33.2.49
Abstract
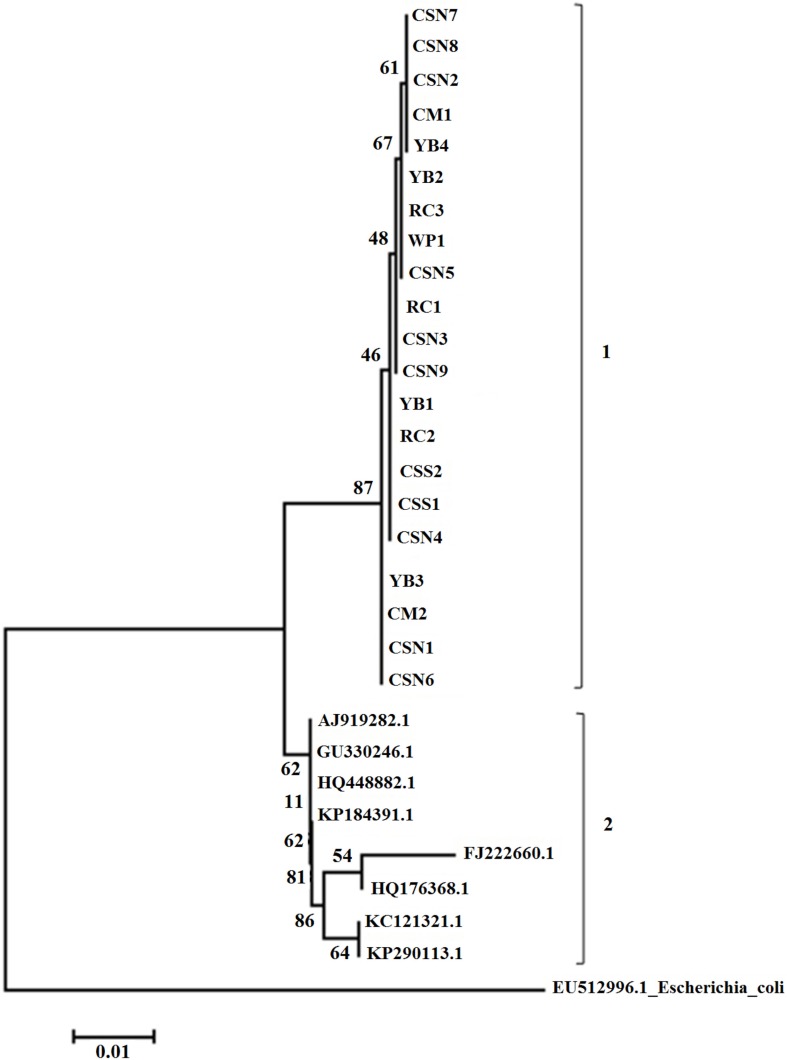
- Turtle-borne Salmonella enterica owns significance as a leading cause in human salmonellosis. The current study aimed to determine the quinolone susceptibility and the genetic characteristics of 21 strains of S. enterica subsp. enterica isolated from pet turtles. Susceptibility of four antimicrobials including nalidixic acid, ciprofloxacin, ofloxacin, and levofloxacin was examined in disk diffusion and MIC tests where the majority of the isolates were susceptible to all tested quinolones. In genetic characterization, none of the isolates were positive for qnr or aac(6')-Ib genes and no any target site mutations could be detected in gyrA, gyrB, and parC quinolone resistance determining regions (QRDR). In addition, neighbor-joining phylogenetic tree derived using gyrA gene sequences exhibited two distinct clads comprising; first, current study isolates, and second, quinolone-resistant isolates of human and animal origin. All results suggest that studied strains of S. enterica subsp. enterica isolated from pet turtles are susceptible to quinolones and genetically more conserved with regards to gyrA gene region.
MeSH Terms
Figure
Reference
-
1. Hui YH, Gorham JR, Murrell KD. Vol. 1; Diseases Caused by Bacteria. Foodborne Disease Handbook. New York: Marcel Dekker Inc.;1994. p. 97–131.2. Barrow PA, Methner U. Salmonella in Domestic Animals. 2nd ed. UK: CABI;2013. p. 136–351.3. Marcus R. New information about pediatric foodborne infections: the view from FoodNet. Curr Opin Pediatr. 2008; 20(1):79–84. PMID: 18197044.
Article4. Bennett SD, Manikonda K, Mungai E, Dewey-Mattia D, Gould LH. Surveillance. 2014. cited 2016 Dec 31. Available from: https://stacks.cdc.gov/view/cdc/23299.5. Kruse H, kirkemo AM, Handeland K. Wildlife as source of zoonotic infections. Emerg Infect Dis. 2004; 10(12):2067–2072. PMID: 15663840.
Article6. Eng SK, Pusparajah P, Ab Mutalib NS, Ser HL, Chan KG, Lee LH. Salmonella: A review on pathogenesis, epidemiology and antibiotic resistance. Front Life Sci. 2015; 8(3):284–293.7. Kownhar H, Shankar EM, Rajan R, Rao UA. Emergence of nalidixic acid-resistant Salmonella enterica serovar Typhi resistant to ciprofloxacin in India. J Med Microbiol. 2007; 56(Pt 1):136–137. PMID: 17172529.
Article8. Souza RB, Ferrari RG, Magnani M, Kottwitz LB, Alcocer I, Tognim MC, Oliveira TC. Ciprofloxacin susceptibility reduction of Salmonella strains isolated from outbreaks. Braz J Microbiol. 2010; 41(2):497–500. PMID: 24031522.
Article9. Carrique-Mas JJ, Papadopoulou C, Evans SJ, Wales A, Teale CJ, Davies RH. Trends in phage types and antimicrobial resistance of Salmonella enterica serovar Enteritidis isolated from animals in Great Britain from 1990 to 2005. Vet Rec. 2008; 162(17):541–546. PMID: 18441349.
Article10. Meakins S, Fisher IS, Berghold C, Gerner-Smidt P, Tschäpe H, Cormican M, Luzzi I, Schneider F, Wannett W, Coia J, Echeita A, Threlfall EJ. Enter-net participants. Antimicrobial drug resistance in human nontyphoidal Salmonella isolates in Europe 2000-2004: a report from the Enter-net International Surveillance Network. Microb Drug Resist. 2008; 14(1):31–35. PMID: 18366323.
Article11. Eaves DJ, Randall L, Gray DT, Buckley A, Woodward MJ, White AP, Piddock LJ. Prevalence of mutations within the quinolone resistance-determining region of gyrA, gyrB, parC, and parE and association with antibiotic resistance in quinolone-resistant Salmonella enterica. Antimicrob Agents Chemother. 2004; 48(10):4012–4015. PMID: 15388468.12. Avsaroglu MD, Helmuth R, Junker E, Hertwig S, Schroeter A, Akcelik M, Bozoglu F, Guerra B. Plasmid-mediated quinolone resistance conferred by qnrS1 in Salmonella enterica serovar Virchow isolated from Turkish food of avian origin. J Antimicrob Chemother. 2007; 60(5):1146–1150. PMID: 17881633.
Article13. Rodríguez-Martínez JM, Cano ME, Velasco C, Martínez-Martínez L, Pascual A. Plasmid-mediated quinolone resistance: an update. J Infect Chemother. 2011; 17(2):149–182. PMID: 20886256.
Article14. Warwick C, Arena PC, Steedman C. Health implications associated with exposure to farmed and wild sea turtles. JRSM Short Rep. 2013; 4(1):8. PMID: 23413410.
Article15. Warwick C, Arena PC, Steedman C, Jessop M. A review of captive exotic animal-linked zoonoses. J Environ Health Res. 2012; 12(1):9–24.16. Shin D-M, Hossain S, Wimalasena S, Heo G-J. Antimicrobial resistance and virulence factors of Edwardsiella tarda isolated from pet turtles. Pak Vet J. 2016; 37(1):85–89.17. Wendt M, Heo GJ. Multilocus sequence typing analysis of Pseudomonas aeruginosa isolated from pet Chinese stripe-necked turtles (Ocadia sinensis). Lab Anim Res. 2016; 32(4):208–216. PMID: 28053614.18. Hossain S, Wimalasena SHMP, Heo G-J. Virulence factors and antimicrobial resistance pattern of Citrobacter freundii isolated from healthy pet turtles and their environment. Asian J Anim Vet Adv. 2017; 12(1):10–16.19. Bosch S, Tauxe RV, Behravesh CB. Turtle-Associated Salmonellosis, United States, 2006-2014. Emerg Infect Dis. 2016; 22(7):1149–1155. PMID: 27315584.
Article20. Cohen ML, Potter M, Pollard R, Feldman RA. Turtle-associated salmonellosis in the United States. Effect of Public Health Action, 1970 to 1976. JAMA. 1980; 243(12):1247–1249. PMID: 7359680.
Article21. Díaz MA, Cooper RK, Cloeckaert A, Siebeling RJ. Plasmid-mediated high-level gentamicin resistance among enteric bacteria isolated from pet turtles in Louisiana. Appl Environ Microbiol. 2006; 72(1):306–312. PMID: 16391058.
Article22. Nowakiewicz A, Ziółkowska G, Zięba P, Stępniewska K, Tokarzewski S. Russian tortoises (Agrionemys horsfieldi) as a potential reservoir for Salmonella spp. Res Vet Sci. 2012; 92(2):187–190. PMID: 21486674.
Article23. Giacopello C, Foti M, Passantino A, Fisichella V, Aleo A, Mammina C. Serotypes and antibiotic susceptibility patterns of Salmonella spp. isolates from spur-thighed tortoise, Testudo graeca illegally introduced in Italy. HVM Bioflux. 2012; 4(2):76–81.24. Bertelloni F, Chemaly M, Cerri D, Gall FL, Ebani VV. Salmonella infection in healthy pet reptiles: Bacteriological isolation and study of some pathogenic characters. Acta Microbiol Immunol Hung. 2016; 63(2):203–216. PMID: 27352973.25. Back DS, Shin GW, Wendt M, Heo GJ. Prevalence of Salmonella spp. in pet turtles and their environment. Lab Anim Res. 2016; 32(3):166–170. PMID: 27729933.26. Bluvias JE, Eckert KL. Marine Turtle Trauma Response Procedures: A Husbandry Manual, Wider Caribbean Sea Turtle Conservation Network (WIDECAST). 2008. p. 11–46.27. CLSI. Performance standards for antimicrobial susceptibility testing: Twenty-fourth informational supplement: CLSI M100-S24. Wayne, USA: Clinical and Laboratory Standards Institute (CLSI);2014.28. Giacopello C, Foti M, Fisichella V, Latella G, Aleo A, Mammina C. Antibiotic resistance in Salmonella isolated from tegus (Tupinambis spp.). J Exot Pet Med. 2012; 21(4):328–331.29. Corrente M, Madio A, Friedrich KG, Greco G, Desario C, Tagliabue S, D'Incau M, Campolo M, Buonavoglia C. Isolation of Salmonella strains from reptile faeces and comparison of different culture media. J Appl Microbiol. 2004; 96(4):709–715. PMID: 15012809.
Article30. Ferrari R, Galiana A, Cremades R, Rodríguez JC, Magnani M, Tognim MC, Oliveira TC, Royo G. Plasmid-mediated quinolone resistance (PMQR) and mutations in the topoisomerase genes of Salmonella enterica strains from Brazil. Braz J Microbiol. 2013; 44(2):651–656. PMID: 24294265.
Article31. Gay K, Robicsek A, Strahilevitz J, Park CH, Jacoby G, Barrett TJ, Medalla F, Chiller TM, Hooper DC. Plasmid-mediated quinolone resistance in non-Typhi serotypes of Salmonella enterica. Clin Infect Dis. 2006; 43(3):297–304. PMID: 16804843.32. Nordmann P, Poirel L. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J Antimicrob Chemother. 2005; 56(3):463–469. PMID: 16020539.
Article33. Kim JH, Cho JK, Kim KS. Prevalence and characterization of plasmid-mediated quinolone resistance genes in Salmonella isolated from poultry in Korea. Avian Pathol. 2013; 42(3):221–229. PMID: 23607509.34. Asai T, Sato C, Masani K, Usui M, Ozawa M, Ogino T, Aoki H, Sawada T, Izumiya H, Watanabe H. Epidemiology of plasmid-mediated quinolone resistance in salmonella enterica serovar typhimurium isolates from food-producing animals in Japan. Gut Pathog. 2010; 2(1):17. PMID: 21138594.
Article35. García-Fernández A, Gallina S, Owczarek S, Dionisi AM, Benedetti I, Decastelli L, Luzzi I. Emergence of Ciprofloxacin-Resistant Salmonella enterica Serovar Typhi in Italy. PLoS One. 2015; 10(6):e0132065. PMID: 26121266.
Article36. Kim SY, Lee SK, Park MS, Na HT. Analysis of the Fluoroquinolone Antibiotic Resistance Mechanism of Salmonella enterica Isolates. J Microbiol Biotechnol. 2016; 26(9):1605–1612. PMID: 27116992.
Article37. Dimitrov T, Dashti AA, Albaksami O, Udo EE, Jadaon MM, Albert MJ. Ciprofloxacin-resistant Salmonella enterica serovar typhi from Kuwait with novel mutations in gyrA and parC genes. J Clin Microbiol. 2009; 47(1):208–211. PMID: 18971360.38. Amarantini C, Satwika D. Molecular phylogeny of Salmonellae: Relationships among Salmonella species determined from gyrA, gyrB, parC, and parE genes. Microbiol Indones. 2015; 9(1):1–8.39. Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. Prevalence in the United States of aac(6')-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob Agents Chemother. 2006; 50(11):3953–3955. PMID: 16954321.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Prevalence of Salmonella spp. in pet turtles and their environment
- Antimicrobial property of lemongrass (Cymbopogon citratus) oil against pathogenic bacteria isolated from pet turtles
- Salmonella enterica subsp. enterica infections in eastern great egrets (Ardea alba modesta)
- Salmonella Serovars from Foodborne and Waterborne Diseases in Korea, 1998-2007: Total Isolates Decreasing Versus Rare Serovars Emerging
- Prevalence of Salmonella enterica and Shiga toxin-producing Escherichia coli in zoo animals from Chile