Endocrinol Metab.
2015 Sep;30(3):381-388. 10.3803/EnM.2015.30.3.381.
Sumoylation of Hes6 Regulates Protein Degradation and Hes1-Mediated Transcription
- Affiliations
-
- 1Department of Brain and Cognitive Sciences, Seoul National University School of Biological Sciences, Seoul, Korea. kyungjin@dgist.ac.kr
- 2Department of Legal Medicine, Korea University College of Medicine, Seoul, Korea.
- KMID: 2407090
- DOI: http://doi.org/10.3803/EnM.2015.30.3.381
Abstract
- BACKGROUND
Hes6 is a transcriptional regulator that induces transcriptional activation by binding to transcription repressor Hes1 and suppressing its activity. Hes6 is controlled by the ubiquitin-proteosome-mediated degradation system. Here we investigated the sumoylation of Hes6 and its functional role in its rhythmic expression.
METHODS
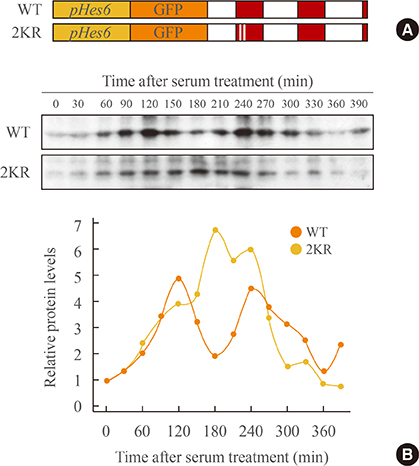
Hes6, SUMO, and ubiquitin were transfected into HeLa cells and the expression pattern was observed by Western blot and immunoprecipitation. To confirm the effect of sumoylation on the rhythmic expression of Hes6, we generated mouse Hes6 promoter-driven GFP-Hes6 fusion constructs and expressed these constructs in NIH 3T3 cells.
RESULTS
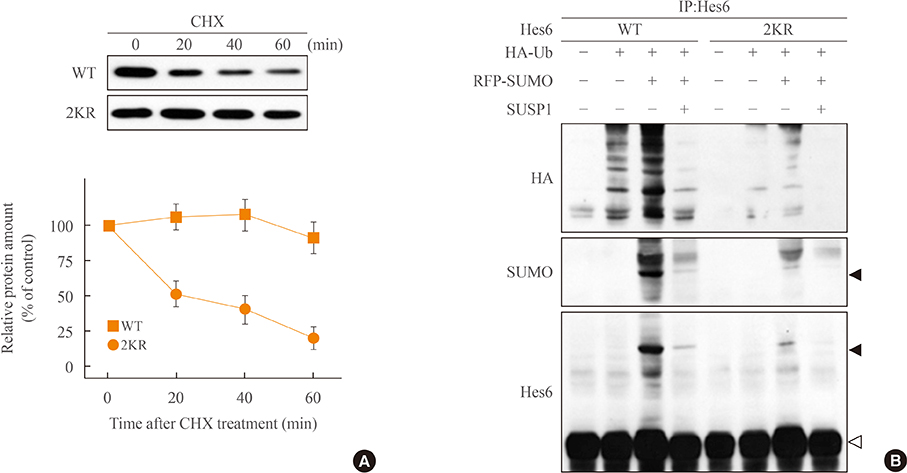
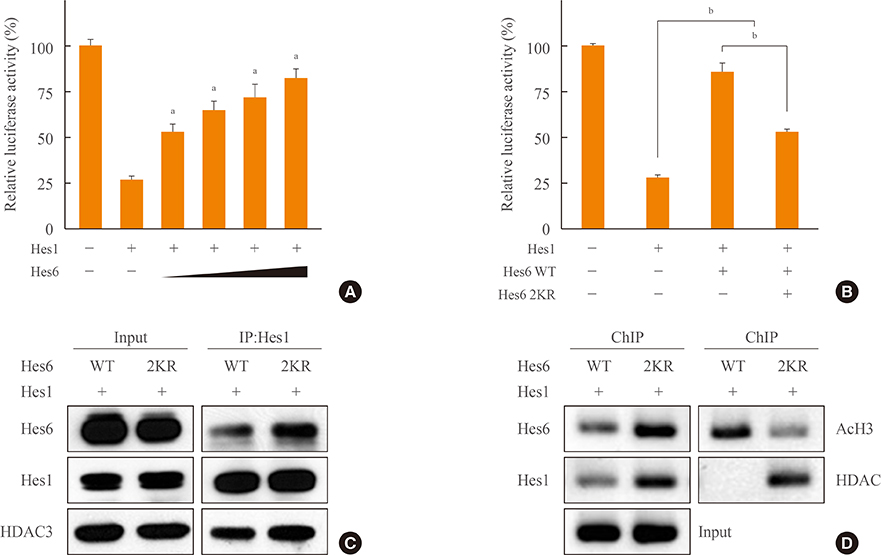
Overexpression of SUMO led to sumoylation of Hes6 at both lysine 27 and 30. Protein stability of Hes6 was decreased by sumoylation. Moreover, expression of a Hes6 sumoylation-defective mutant, the 2KR (K27/30R) mutant, or co-expression of SUMO protease SUSP1 with native Hes6, strongly reduced ubiquitination. In addition, sumoylation was associated with both the rhythmic expression and transcriptional regulation of Hes6. Wild type Hes6 showed oscillatory expression with about 2-hour periodicity, whereas the 2KR mutant displayed a longer period. Furthermore, sumoylation of Hes6 derepressed Hes1-induced transcriptional repression.
CONCLUSION
Hes6 sumoylation plays an important role in the regulation of its stability and Hes1-mediated transcription. These results suggest that sumoylation may be crucial for rhythmic expression of Hes6 and downstream target genes.
Keyword
MeSH Terms
Figure
Reference
-
1. Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002; 3:517–530.2. Ross SE, Greenberg ME, Stiles CD. Basic helix-loop-helix factors in cortical development. Neuron. 2003; 39:13–25.3. Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development. 2007; 134:1243–1251.4. Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 1992; 6:2620–2634.5. Chen H, Thiagalingam A, Chopra H, Borges MW, Feder JN, Nelkin BD, et al. Conservation of the Drosophila lateral inhibition pathway in human lung cancer: a hairy-related protein (HES-1) directly represses achaete-scute homolog-1 expression. Proc Natl Acad Sci U S A. 1997; 94:5355–5360.6. Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995; 377:355–358.7. Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999; 18:2196–2207.8. Bae S, Bessho Y, Hojo M, Kageyama R. The bHLH gene Hes6, an inhibitor of Hes1, promotes neuronal differentiation. Development. 2000; 127:2933–2943.9. Koyano-Nakagawa N, Kim J, Anderson D, Kintner C. Hes6 acts in a positive feedback loop with the neurogenins to promote neuronal differentiation. Development. 2000; 127:4203–4216.10. Harima Y, Imayoshi I, Shimojo H, Kobayashi T, Kageyama R. The roles and mechanism of ultradian oscillatory expression of the mouse Hes genes. Semin Cell Dev Biol. 2014; 34:85–90.11. Pascoal S, Carvalho CR, Rodriguez-Leon J, Delfini MC, Duprez D, Thorsteinsdottir S, et al. A molecular clock operates during chick autopod proximal-distal outgrowth. J Mol Biol. 2007; 368:303–309.12. Schwendinger-Schreck J, Kang Y, Holley SA. Modeling the zebrafish segmentation clock's gene regulatory network constrained by expression data suggests evolutionary transitions between oscillating and nonoscillating transcription. Genetics. 2014; 197:725–738.13. Masamizu Y, Ohtsuka T, Takashima Y, Nagahara H, Takenaka Y, Yoshikawa K, et al. Real-time imaging of the somite segmentation clock: revelation of unstable oscillators in the individual presomitic mesoderm cells. Proc Natl Acad Sci U S A. 2006; 103:1313–1318.14. Shimojo H, Ohtsuka T, Kageyama R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron. 2008; 58:52–64.15. Hirata H, Yoshiura S, Ohtsuka T, Bessho Y, Harada T, Yoshikawa K, et al. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science. 2002; 298:840–843.16. Hirata H, Bessho Y, Kokubu H, Masamizu Y, Yamada S, Lewis J, et al. Instability of Hes7 protein is crucial for the somite segmentation clock. Nat Genet. 2004; 36:750–754.17. Bessho Y, Sakata R, Komatsu S, Shiota K, Yamada S, Kageyama R. Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev. 2001; 15:2642–2647.18. Takebayashi K, Sasai Y, Sakai Y, Watanabe T, Nakanishi S, Kageyama R. Structure, chromosomal locus, and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-1. Negative autoregulation through the multiple N box elements. J Biol Chem. 1994; 269:5150–5156.19. Kang SA, Seol JH, Kim J. The conserved WRPW motif of Hes6 mediates proteasomal degradation. Biochem Biophys Res Commun. 2005; 332:33–36.20. Hay RT. SUMO: a history of modification. Mol Cell. 2005; 18:1–12.21. Chiou HY, Liu SY, Lin CH, Lee EH. Hes-1 SUMOylation by protein inhibitor of activated STAT1 enhances the suppressing effect of Hes-1 on GADD45alpha expression to increase cell survival. J Biomed Sci. 2014; 21:53.22. Denuc A, Marfany G. SUMO and ubiquitin paths converge. Biochem Soc Trans. 2010; 38(Pt 1):34–39.23. Schimmel J, Larsen KM, Matic I, van Hagen M, Cox J, Mann M, et al. The ubiquitin-proteasome system is a key component of the SUMO-2/3 cycle. Mol Cell Proteomics. 2008; 7:2107–2122.24. Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998; 2:233–239.25. Lee J, Lee Y, Lee MJ, Park E, Kang SH, Chung CH, et al. Dual modification of BMAL1 by SUMO2/3 and ubiquitin promotes circadian activation of the CLOCK/BMAL1 complex. Mol Cell Biol. 2008; 28:6056–6065.26. Cardone L, Hirayama J, Giordano F, Tamaru T, Palvimo JJ, Sassone-Corsi P. Circadian clock control by SUMOylation of BMAL1. Science. 2005; 309:1390–1394.27. Kageyama R, Ohtsuka T, Kobayashi T. Roles of Hes genes in neural development. Dev Growth Differ. 2008; 50:Suppl 1. S97–S103.28. Lee YJ, Han DH, Pak YK, Cho SH. Circadian regulation of low density lipoprotein receptor promoter activity by CLOCK/BMAL1, Hes1 and Hes6. Exp Mol Med. 2012; 44:642–652.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Circadian regulation of low density lipoprotein receptor promoter activity by CLOCK/BMAL1, Hes1 and Hes6
- SUMO Proteins are not Involved in TGF-beta1-induced, Smad3/4-mediated Germline alpha Transcription, but PIASy Suppresses it in CH12F3-2A B Cells
- Endoplasmic Reticulum Stress-Mediated p62 Downregulation Inhibits Apoptosis via c-Jun Upregulation
- Regulation of Protein Degradation by Proteasomes in Cancer
- Dyrk1A Positively Stimulates ASK1-JNK Signaling Pathway during Apoptotic Cell Death