Immune Netw.
2017 Oct;17(5):307-316. 10.4110/in.2017.17.5.307.
Semi-Functional Quantitative Flow Cytometry Assay for Lymphocytic Choriomeningitis Virus Titration
- Affiliations
-
- 1Department of Biochemistry, College of Life Science & Biotechnology, Yonsei University Seoul 03722, Korea. sjha@yonsei.ac.kr
- KMID: 2400636
- DOI: http://doi.org/10.4110/in.2017.17.5.307
Abstract
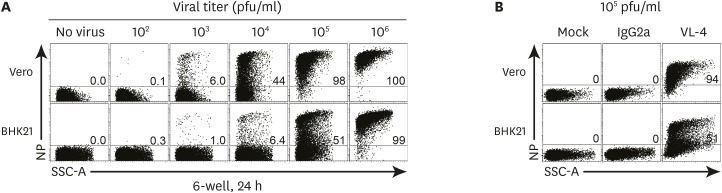
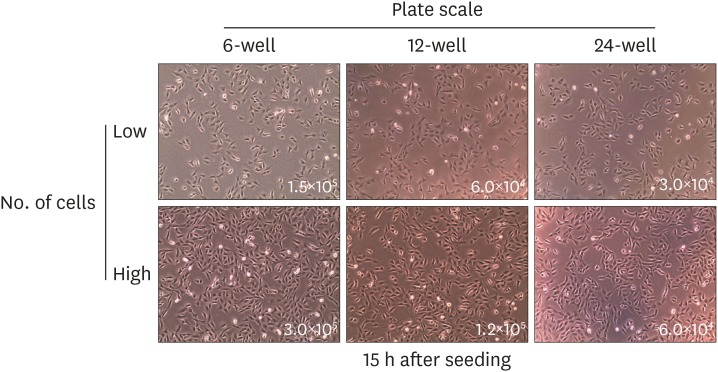
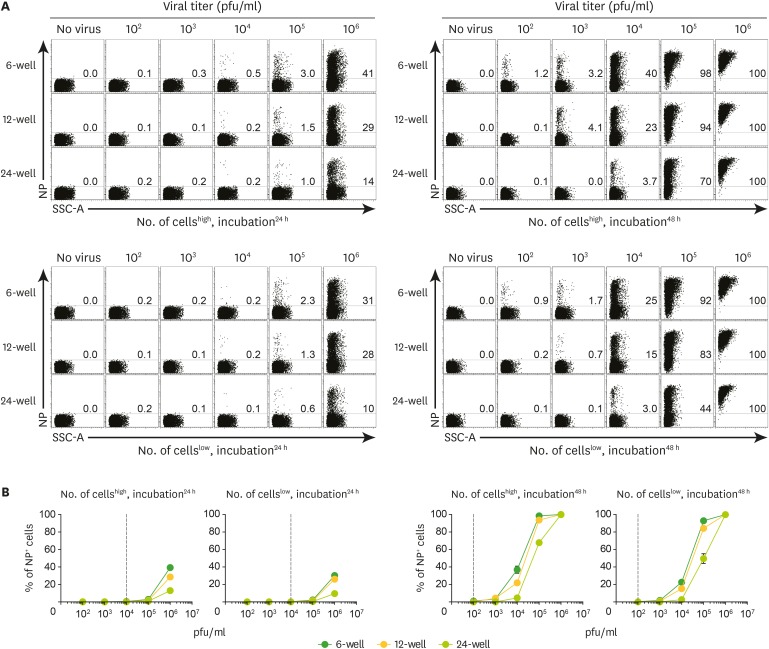
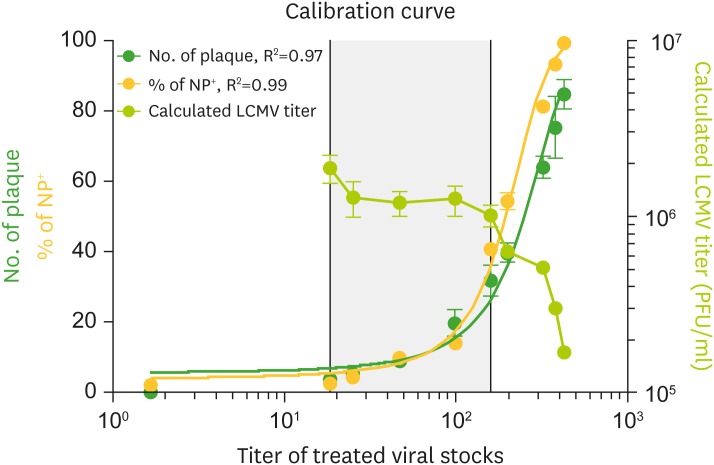
- Quantitative PCR and plaque assay are powerful virological techniques used to measure the load of defective or infectious virus in mouse and human. However, these methods display limitations such as cross contamination and long run-time. Here, we describe a novel technique termed as semi-functional quantitative flow cytometry (SFQF) for the accurate estimation of the quantity of infectious lymphocytic choriomeningitis virus (LCMV). LCMV titration method using flow cytometry was previously developed but has technical shortcomings, owing to the less optimized parameters such as cell overgrowth, plate scale, and detection threshold. Therefore, we first established optimized conditions for SFQF assay using LCMV nucleoprotein (NP)-specific antibody to evaluate the threshold of the virus detection range in the plaque assay. We subsequently demonstrated that the optimization of the method increased the sensitivity of virus detection. We revealed several new advantages of SFQF assay, which overcomes some of the previously contentious points, and established an upgraded version of the previously reported flow cytometric titration assay. This method extends the detection scale to the level of single cell, allowing extension of its application for in vivo detection of infected cells and their phenotypic analysis. Thus, SFQF assay may serve as an alternative analytical tool for ensuring the reliability of LCMV titration and can be used with other types of viruses using target-specific antibodies.
Keyword
MeSH Terms
Figure
Reference
-
1. Zhou X, Ramachandran S, Mann M, Popkin DL. Role of lymphocytic choriomeningitis virus (LCMV) in understanding viral immunology: past, present and future. Viruses. 2012; 4:2650–2669. PMID: 23202498.
Article2. Salvato MS, Shimomaye EM. The completed sequence of lymphocytic choriomeningitis virus reveals a unique RNA structure and a gene for a zinc finger protein. Virology. 1989; 173:1–10. PMID: 2510401.
Article3. Pinschewer DD, Perez M, de la Torre JC. Role of the virus nucleoprotein in the regulation of lymphocytic choriomeningitis virus transcription and RNA replication. J Virol. 2003; 77:3882–3887. PMID: 12610166.
Article4. Darbre S, Johnson S, Kallert S, Lambert PH, Siegrist CA, Pinschewer DD. The nucleoprotein is required for lymphocytic choriomeningitis virus-based vaccine vector immunogenicity. J Virol. 2015; 89:11734–11738. PMID: 26355095.
Article5. Marcus PI, Carver DH. Hemadsorption-negative plaque test: new assay for rubella virus revealing a unique interference. Science. 1965; 149:983–986. PMID: 4283964.
Article6. Cooper PD. The plaque assay of animal viruses. Adv Virus Res. 1961; 8:319–378. PMID: 13881155.
Article7. Sedwick WD, Wiktor TJ. Reproducible plaquing system for rabies, lymphocytic choriomeningitis,k and other ribonucleic acid viruses in BHK-21-13S agarose suspensions. J Virol. 1967; 1:1224–1226. PMID: 4987176.8. Wainwright S, Mims CA. Plaque assay for lymphocytic choriomeningitis virus based on hemadsorption interference. J Virol. 1967; 1:1091–1092. PMID: 4987174.
Article9. McCausland MM, Crotty S. Quantitative PCR technique for detecting lymphocytic choriomeningitis virus in vivo. J Virol Methods. 2008; 147:167–176. PMID: 17920702.10. Cordey S, Sahli R, Moraz ML, Estrade C, Morandi L, Cherpillod P, Charrel RN, Kunz S, Kaiser L. Analytical validation of a lymphocytic choriomeningitis virus real-time RT-PCR assay. J Virol Methods. 2011; 177:118–122. PMID: 21763351.
Article11. Welsh RM, Pfau CJ. Determinants of lymphocytic choriomeningitis interference. J Gen Virol. 1972; 14:177–187. PMID: 4622135.
Article12. Welsh RM Jr, Buchmeier MJ. Protein analysis of defective interfering lymphocytic choriomeningitis virus and persistently infected cells. Virology. 1979; 96:503–515. PMID: 380148.
Article13. Welsh RM, Oldstone MB. Inhibition of immunologic injury of cultured cells infected with lymphocytic choriomeningitis virus: role of defective interfering virus in regulating viral antigenic expression. J Exp Med. 1977; 145:1449–1468. PMID: 301173.
Article14. Sheĭnbergas MM, Vorob’eva ZN. Titration of antibodies to lymphocytic choriomeningitis virus by the method of indirect immunofluorescence. Vopr Virusol. 1975; 357–360. PMID: 1099805.15. Battegay M, Cooper S, Althage A, Bänziger J, Hengartner H, Zinkernagel RM. Quantification of lymphocytic choriomeningitis virus with an immunological focus assay in 24- or 96-well plates. J Virol Methods. 1991; 33:191–198. PMID: 1939506.
Article16. Battegay M. Quantification of lymphocytic choriomeningitis virus with an immunological focus assay in 24 well plates. ALTEX. 1993; 10:6–14. PMID: 11178349.17. Korns Johnson D, Homann D. Accelerated and improved quantification of lymphocytic choriomeningitis virus (LCMV) titers by flow cytometry. PLoS One. 2012; 7:e37337. PMID: 22615984.
Article18. Dutko FJ, Oldstone MB. Genomic and biological variation among commonly used lymphocytic choriomeningitis virus strains. J Gen Virol. 1983; 64:1689–1698. PMID: 6875516.
Article19. Ahmed R, Salmi A, Butler LD, Chiller JM, Oldstone MB. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J Exp Med. 1984; 160:521–540. PMID: 6332167.
Article20. Cao W, Henry MD, Borrow P, Yamada H, Elder JH, Ravkov EV, Nichol ST, Compans RW, Campbell KP, Oldstone MB. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science. 1998; 282:2079–2081. PMID: 9851928.21. Welsh RM, Seedhom MO. Lymphocytic choriomeningitis virus (LCMV): propagation, quantitation, and storage. Curr Protoc Microbiol. 2008; Chapter 15:Unit 15A.1.
Article22. Bocharov G, Ludewig B, Bertoletti A, Klenerman P, Junt T, Krebs P, Luzyanina T, Fraser C, Anderson RM. Underwhelming the immune response: effect of slow virus growth on CD8+-T-lymphocyte responses. J Virol. 2004; 78:2247–2254. PMID: 14963121.23. Ammerman NC, Beier-Sexton M, Azad AF. Growth and maintenance of Vero cell lines. Curr Protoc Microbiol . Appendix. 2008; Appendix 4:Appendix 4E.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Comparative Study of the Standard Plaque Assay with Solid-overlay and Immunofocus Assay for Varicella-zoster Virus Titration
- Extrinsic Acquisition of CD80 by Antigen-Specific CD8⺠T Cells Regulates Their Recall Immune Responses to Acute Viral Infection
- The Multifaceted Roles of NK Cells in the Context of Murine Cytomegalovirus and Lymphocytic Choriomeningitis Virus Infections
- Re-defining T-Cell Exhaustion: Subset, Function, and Regulation
- Metabolic Reprogramming by the Excessive AMPK Activation Exacerbates Antigen-Specific Memory CD8⺠T Cell Differentiation after Acute Lymphocytic Choriomeningitis Virus Infection