Tuberc Respir Dis.
2017 Jul;80(3):255-264. 10.4046/trd.2017.80.3.255.
The Role of N-Acetyl Transferases on Isoniazid Resistance from Mycobacterium tuberculosis and Human: An In Silico Approach
- Affiliations
-
- 1Department of Biomedical Informatics, National Institute for Research in Tuberculosis (NIRT), Indian Council of Medical Research (ICMR), Chennai, India. nusrathunissa@gmail.com
- KMID: 2385040
- DOI: http://doi.org/10.4046/trd.2017.80.3.255
Abstract
- BACKGROUND
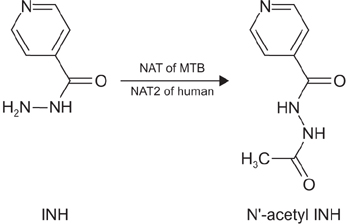
N-acetyl transferase (NAT) inactivates the pro-drug isoniazid (INH) to N-acetyl INH through a process of acetylation, and confers low-level resistance to INH in Mycobacterium tuberculosis (MTB). Similar to NAT of MTB, NAT2 in humans performs the same function of acetylation. Rapid acetylators, may not respond to INH treatment efficiently, and could be a potential risk factor, for the development of INH resistance in humans.
METHODS
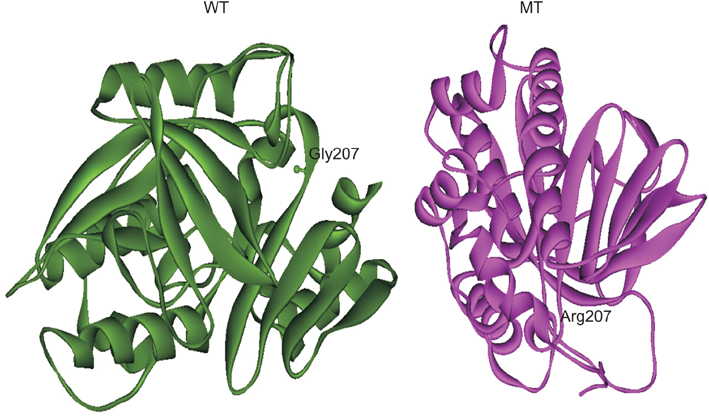
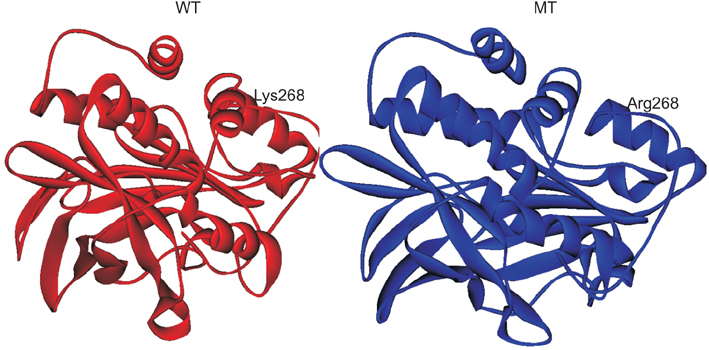
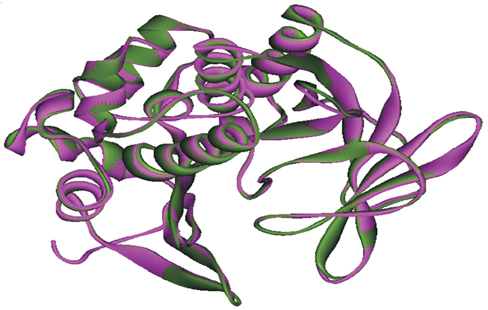
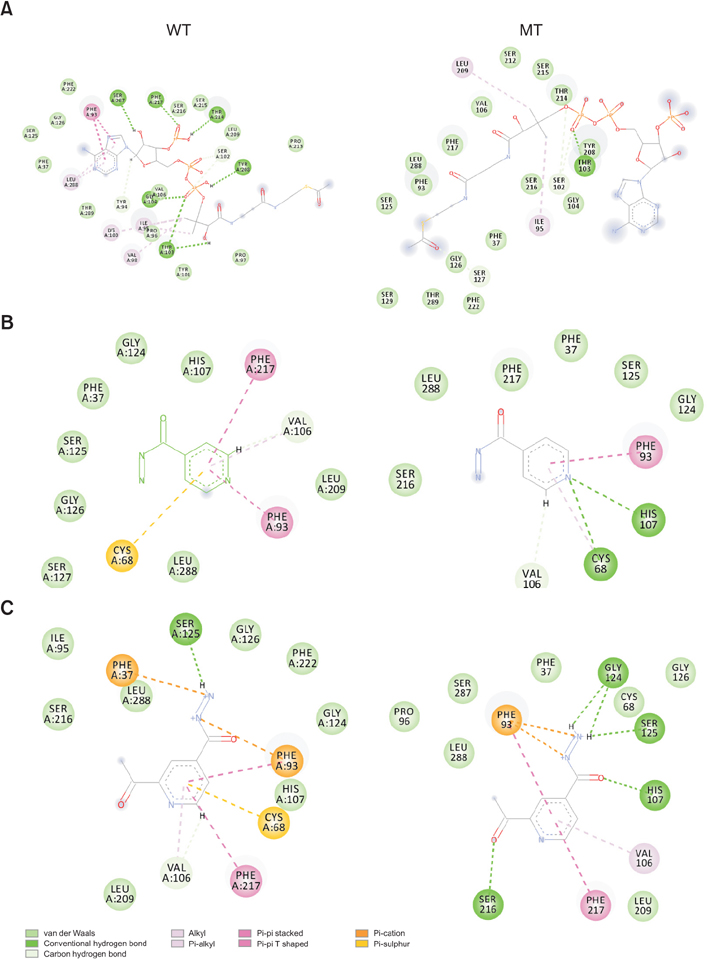
To understand the contribution of NAT of MTB and NAT2 of humans in developing INH resistance using in silico approaches, in this study, the wild type (WT) and mutant (MT)-NATs of MTB, and humans, were modeled and docked, with substrates and product (acetyl CoA, INH, and acetyl INH). The MT models were built, using templates 4BGF of MTB, and 2PFR of humans.
RESULTS
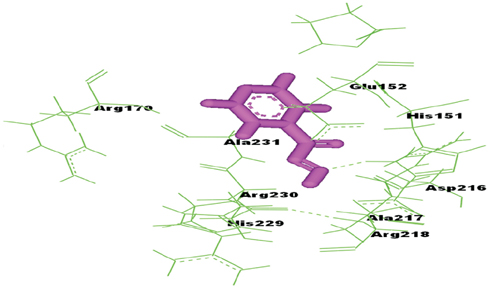
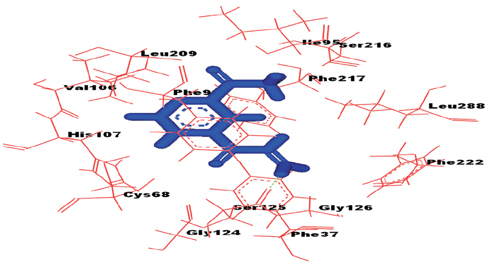
On the basis of docking results of MTB-NAT, it can be suggested that in comparison to the WT, binding affinity of MT-G207R, was found to be lower with acetyl CoA, and higher with acetyl-INH and INH. In case of MT-NAT2 from humans, the pattern of score with respect to acetyl CoA and acetyl-INH, was similar to MT-NAT of MTB, but revealed a decrease in INH score.
CONCLUSION
In MTB, MT-NAT revealed high affinity towards acetyl-INH, which can be interpreted as increased formation of acetyl-INH, and therefore, may lead to INH resistance through inactivation of INH. Similarly, in MT-NAT2 (rapid acetylators), acetylation occurs rapidly, serving as a possible risk factor for developing INH resistance in humans.
Keyword
MeSH Terms
Figure
Reference
-
1. World Health Organization. Anti-tuberculosis drug resistance in the world. Fourth Global Report. The WHO/IAUTLD Global Project on Anti-tuberculosis Drug Resistance Surveillance, 2002-2007. Geneva: World Health Organization;2008.2. Winder F. Catalase and peroxidase in mycobacteria: possible relationship to the mode of action of isoniazid. Am Rev Respir Dis. 1960; 81:68–78.3. Zhang Y, Heym B, Allen B, Young D, Cole S. The catalaseperoxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992; 358:591–593.4. Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um KS, Wilson T, et al. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994; 263:227–230.5. Ramaswamy S, Musser JM. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis. 1998; 79:3–29.6. Payton M, Auty R, Delgoda R, Everett M, Sim E. Cloning and characterization of arylamine N-acetyltransferase genes from Mycobacterium smegmatis and Mycobacterium tuberculosis: increased expression results in isoniazid resistance. J Bacteriol. 1999; 181:1343–1347.7. Upton AM, Mushtaq A, Victor TC, Sampson SL, Sandy J, Smith DM, et al. Arylamine N-acetyltransferase of Mycobacterium tuberculosis is a polymorphic enzyme and a site of isoniazid metabolism. Mol Microbiol. 2001; 42:309–317.8. Bhakta S, Besra GS, Upton AM, Parish T, Sholto-Douglas-Vernon C, Gibson KJ, et al. Arylamine N-acetyltransferase is required for synthesis of mycolic acids and complex lipids in Mycobacterium bovis BCG and represents a novel drug target. J Exp Med. 2004; 199:1191–1199.9. Evans DA, Manley KA, Mc KV. Genetic control of isoniazid metabolism in man. Br Med J. 1960; 2:485–491.10. Hickman D, Palamanda JR, Unadkat JD, Sim E. Enzyme kinetic properties of human recombinant arylamine N-acetyltransferase 2 allotypic variants expressed in Escherichia coli. Biochem Pharmacol. 1995; 50:697–703.11. Parkin DP, Vandenplas S, Botha FJ, Vandenplas ML, Seifart HI, van Helden PD, et al. Trimodality of isoniazid elimination: phenotype and genotype in patients with tuberculosis. Am J Respir Crit Care Med. 1997; 155:1717–1722.12. Vatsis KP, Weber WW, Bell DA, Dupret JM, Evans DA, Grant DM, et al. Nomenclature for N-acetyltransferases. Pharmacogenetics. 1995; 5:1–17.13. Kinzig-Schippers M, Tomalik-Scharte D, Jetter A, Scheidel B, Jakob V, Rodamer M, et al. Should we use N-acetyltransferase type 2 genotyping to personalize isoniazid doses? Antimicrob Agents Chemother. 2005; 49:1733–1738.14. Azuma J, Ohno M, Kubota R, Yokota S, Nagai T, Tsuyuguchi K, et al. NAT2 genotype guided regimen reduces isoniazid-induced liver injury and early treatment failure in the 6-month four-drug standard treatment of tuberculosis: a randomized controlled trial for pharmacogenetics-based therapy. Eur J Clin Pharmacol. 2013; 69:1091–1101.15. Huang YS, Chern HD, Su WJ, Wu JC, Lai SL, Yang SY, et al. Polymorphism of the N-acetyltransferase 2 gene as a susceptibility risk factor for antituberculosis drug-induced hepatitis. Hepatology. 2002; 35:883–889.16. Ohno M, Yamaguchi I, Yamamoto I, Fukuda T, Yokota S, Maekura R, et al. Slow N-acetyltransferase 2 genotype affects the incidence of isoniazid and rifampicin-induced hepatotoxicity. Int J Tuberc Lung Dis. 2000; 4:256–261.17. Saukkonen JJ, Cohn DL, Jasmer RM, Schenker S, Jereb JA, Nolan CM, et al. An official ATS statement: hepatotoxicity of antituberculosis therapy. Am J Respir Crit Care Med. 2006; 174:935–952.18. Unissa AN, Subbian S, Hanna LE, Selvakumar N. Overview on mechanisms of isoniazid action and resistance in Mycobacterium tuberculosis. Infect Genet Evol. 2016; 45:474–492.19. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25:3389–3402.20. Abuhammad A, Lowe ED, McDonough MA, Shaw Stewart PD, Kolek SA, Sim E, et al. Structure of arylamine N-acetyltransferase from Mycobacterium tuberculosis determined by cross-seeding with the homologous protein from M. marinum: triumph over adversity. Acta Crystallogr D Biol Crystallogr. 2013; 69(Pt 8):1433–1446.21. Wu H, Dombrovsky L, Tempel W, Martin F, Loppnau P, Goodfellow GH, et al. Structural basis of substrate-binding specificity of human arylamine N-acetyltransferases. J Biol Chem. 2007; 282:30189–30197.22. Sali A. MODELLER: implementing 3D protein modeling. mc2, Vol. 2. San Diego: Molecular Simulations Inc.;1995.23. Lovell SC, Davis IW, Arendall WB 3rd, de Bakker PI, Word JM, Prisant MG, et al. Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins. 2003; 50:437–450.24. Krissinel E, Henrick K. Protein structure comparison in 3D based on secondary structure matching (PDBeFold) followed by Cα alignment, scored by a new structural similarity function. In : Proceedings of the 5th International Conference on Molecular Structural Biology; 2003 Sep 3-7; Vienna, Austria. p. 88.25. Advanced Chemistry Development. ACD/ChemSketch, ver. 10.0. Toronto: Advanced Chemistry Development;2006.26. Jones G, Willett P, Glen RC. Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J Mol Biol. 1995; 245:43–53.27. Accelrys Inc. Discovery studio, ver. 2. San Diego: Accelrys Inc.;2007.28. Abuhammad AM, Lowe ED, Fullam E, Noble M, Garman EF, Sim E. Probing the architecture of the Mycobacterium marinum arylamine N-acetyltransferase active site. Protein Cell. 2010; 1:384–392.29. Fullam E, Westwood IM, Anderton MC, Lowe ED, Sim E, Noble ME. Divergence of cofactor recognition across evolution: coenzyme A binding in a prokaryotic arylamine Nacetyltransferase. J Mol Biol. 2008; 375:178–191.30. Sandy J, Mushtaq A, Kawamura A, Sinclair J, Sim E, Noble M. The structure of arylamine N-acetyltransferase from Mycobacterium smegmatis: an enzyme which inactivates the antitubercular drug, isoniazid. J Mol Biol. 2002; 318:1071–1083.31. Dassault Systemes. BIOVIA, Discovery Studio Modeling Environment. Release 4.5. San Diego: Dassault Systemes;2015.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Understanding Rifampicin Resistance in Tuberculosis through a Computational Approach
- The Relationship between Isoniazid Resistance and 463 CodonMutation of katG Gne in Mycobacterium Tuberculosis
- The Drug Resistance of Mycobacterium tuberculosis in Seongnam
- Molecular analysis of katG gene mutations in strains of Mycobacterium tuberculosis from Korea
- Development of Oligonucleotide Chip for Detection of Drug-Resistant Mycobacterium Tuberculosis