Obstet Gynecol Sci.
2016 Nov;59(6):444-453. 10.5468/ogs.2016.59.6.444.
Quantitative fluorescent polymerase chain reaction for rapid prenatal diagnosis of fetal aneuploidies in chorionic villus sampling in a single institution
- Affiliations
-
- 1Department of Obstetrics and Gynecology, Hankook General Hospital, Jeju, Korea.
- 2Department of Obstetrics and Gynecology, Cheil General Hospital and Women's Healthcare Center, Seoul, Korea. sabi0515@hanmail.net
- 3Laboratory of Medical Genetics, Dankook University College of Medicine, Seoul, Korea.
- KMID: 2378568
- DOI: http://doi.org/10.5468/ogs.2016.59.6.444
Abstract
OBJECTIVE
To validate quantitative fluorescent polymerase chain reaction (QF-PCR) via chorionic villus sampling (CVS) for the diagnosis of fetal aneuploidies.
METHODS
We retrospectively reviewed the medical records of consecutive pregnant women who had undergone CVS at Cheil General Hospital between December 2009 and June 2014. Only cases with reported QF-PCR before long-term culture (LTC) for conventional cytogenetic analysis were included, and the results of these two methods were compared.
RESULTS
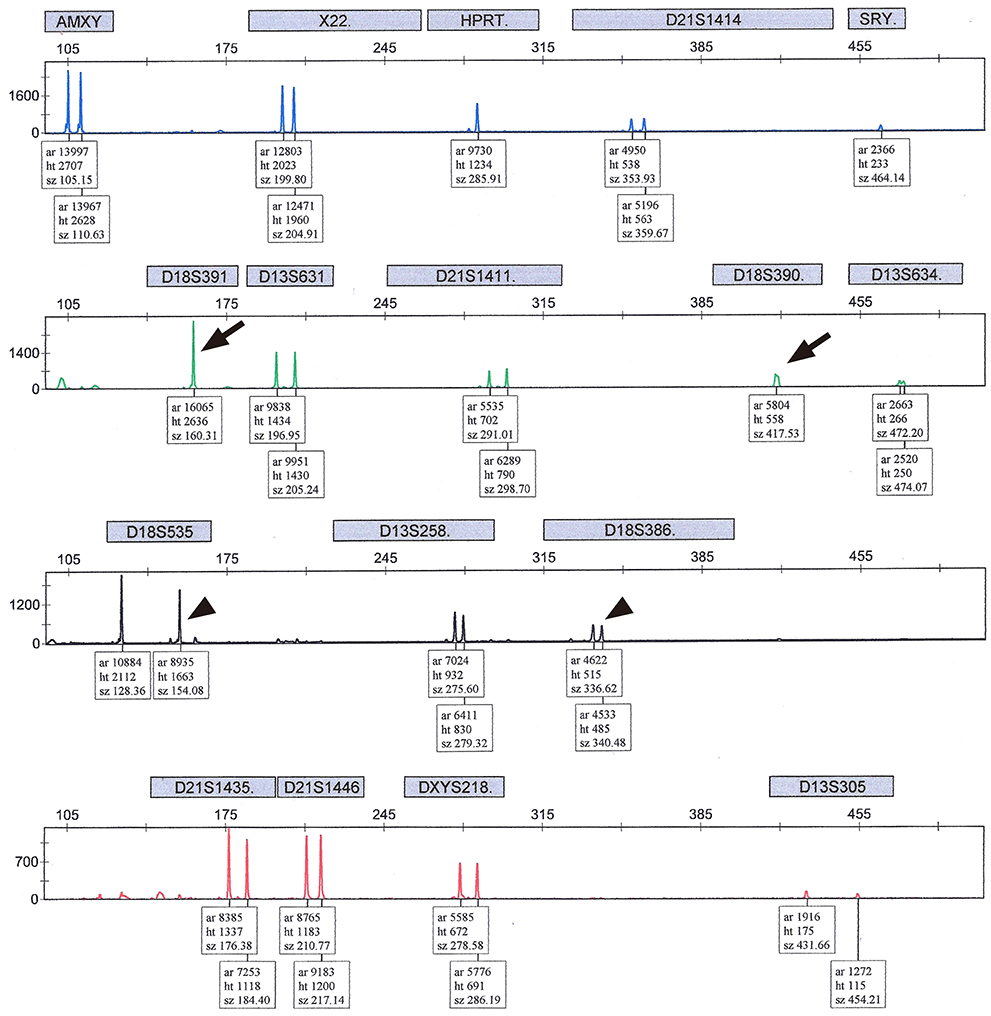
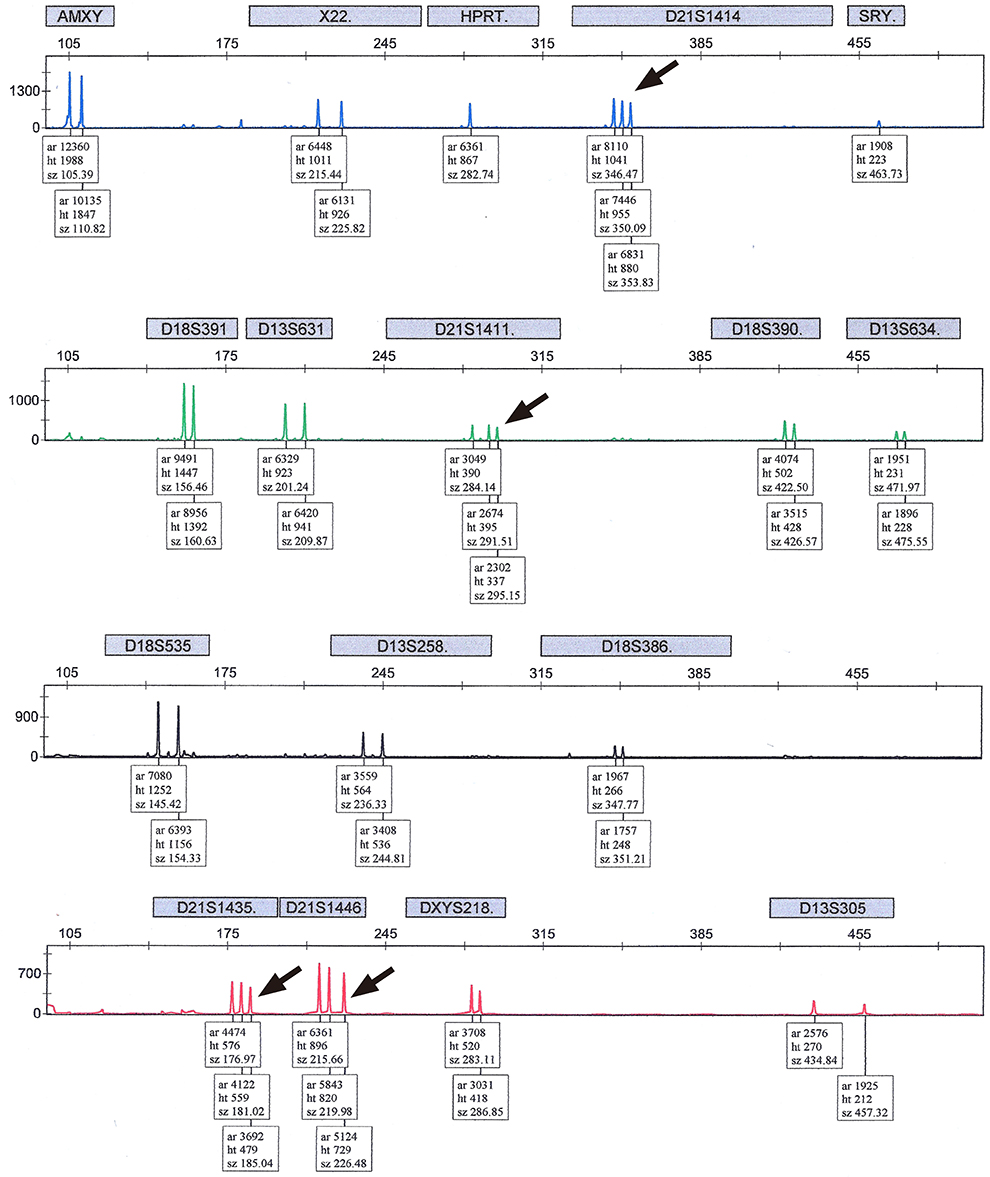
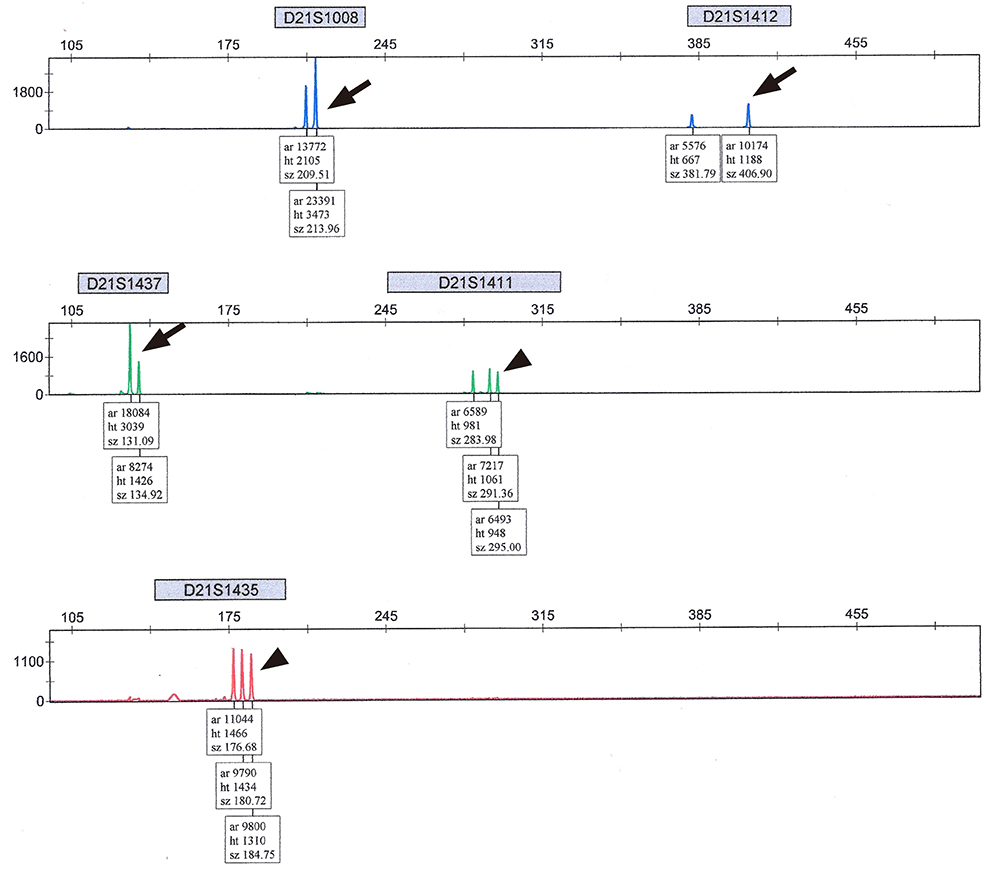
A total of 383 pregnant women underwent QF-PCR and LTC via CVS during the study period and 403 CVS specimens were collected. The indications of CVS were as follows: abnormal first-trimester ultrasonographic findings, including increased fetal nuchal translucency (85.1%), advanced maternal age (6.8%), previous history of fetal anomalies (4.2%), and positive dual test results for trisomy 21 (3.9%). The results of QF-PCR via CVS were as follows: 76 (18.9%) cases were identified as trisomy 21 (36 cases), 18 (33 cases), or 13 (seven cases), and 4 (1.0%) cases were suspected to be mosaicism. All results of common autosomal trisomies by QF-PCR were consistent with those of LTC and there were no false-positive findings. Four cases suspected as mosaicism in QF-PCR were confirmed as non-mosaic trisomies of trisomy 21 (one case) or trisomy 18 (three cases) in LTC.
CONCLUSION
QF-PCR via CVS has the advantage of rapid prenatal screening at an earlier stage of pregnancy for common chromosomal trisomies and thus can reduce the anxiety of parents. In particular, it can be helpful for pregnant women with increased fetal nuchal translucency or abnormal first-trimester ultrasonographic findings.
MeSH Terms
-
Aneuploidy*
Anxiety
Chorion*
Chorionic Villi Sampling*
Chorionic Villi*
Cytogenetic Analysis
Diagnosis
Down Syndrome
Female
Fluorescence
Hospitals, General
Humans
Maternal Age
Medical Records
Mosaicism
Nuchal Translucency Measurement
Parents
Polymerase Chain Reaction*
Pregnancy
Pregnant Women
Prenatal Diagnosis*
Retrospective Studies
Trisomy
Figure
Reference
-
1. Mansfield ES. Diagnosis of Down syndrome and other aneuploidies using quantitative polymerase chain reaction and small tandem repeat polymorphisms. Hum Mol Genet. 1993; 2:43–50.2. Mann K, Donaghue C, Fox SP, Docherty Z, Ogilvie CM. Strategies for the rapid prenatal diagnosis of chromosome aneuploidy. Eur J Hum Genet. 2004; 12:907–915.3. Mann K, Fox SP, Abbs SJ, Yau SC, Scriven PN, Docherty Z, et al. Development and implementation of a new rapid aneuploidy diagnostic service within the UK National Health Service and implications for the future of prenatal diagnosis. Lancet. 2001; 358:1057–1061.4. Pertl B, Kopp S, Kroisel PM, Tului L, Brambati B, Adinolfi M. Rapid detection of chromosome aneuploidies by quantitative fluorescence PCR: first application on 247 chorionic villus samples. J Med Genet. 1999; 36:300–303.5. Levett LJ, Liddle S, Meredith R. A large-scale evaluation of amnio-PCR for the rapid prenatal diagnosis of fetal trisomy. Ultrasound Obstet Gynecol. 2001; 17:115–118.6. Allen SK, Luharia A, Gould CP, MacDonald F, Larkins S, Davison EV. Rapid prenatal diagnosis of common trisomies: discordant results between QF-PCR analysis and karyotype analysis on long-term culture for a case of trisomy 18 detected in CVS. Prenat Diagn. 2006; 26:1160–1167.7. Waters JJ, Mann K, Grimsley L, Ogilvie CM, Donaghue C, Staples L, et al. Complete discrepancy between QF-PCR analysis of uncultured villi and karyotyping of cultured cells in the prenatal diagnosis of trisomy 21 in three CVS. Prenat Diagn. 2007; 27:332–339.8. Waters JJ, Walsh S, Levett LJ, Liddle S, Akinfenwa Y. Complete discrepancy between abnormal fetal karyotypes predicted by QF-PCR rapid testing and karyotyped cultured cells in a first-trimester CVS. Prenat Diagn. 2006; 26:892–897.9. Ogilvie C, Mann K. Discrepancies between direct QF-PCR of chorionic villi and karyotype analysis of chorionic mesenchyme cells. Prenat Diagn. 2012; 32:501–502.10. Chung JH, Yang JH, Song MJ, Cho JY, Lee YH, Park SY, et al. The distribution of fetal nuchal translucency thickness in normal Korean fetuses. J Korean Med Sci. 2004; 19:32–36.11. Grati FR, Malvestiti F, Grimi B, Gaetani E, Di Meco AM, Trotta A, et al. QF-PCR as a substitute for karyotyping of cytotrophoblast for the analysis of chorionic villi: advantages and limitations from a cytogenetic retrospective audit of 44,727 first-trimester prenatal diagnoses. Prenat Diagn. 2013; 33:502–508.12. Mann K, Ogilvie CM. QF-PCR: application, overview and review of the literature. Prenat Diagn. 2012; 32:309–314.13. Cirigliano V, Voglino G, Ordonez E, Marongiu A, Paz Canadas M, Ejarque M, et al. Rapid prenatal diagnosis of common chromosome aneuploidies by QF-PCR, results of 9 years of clinical experience. Prenat Diagn. 2009; 29:40–49.14. Cirigliano V, Voglino G, Canadas MP, Marongiu A, Ejarque M, Ordonez E, et al. Rapid prenatal diagnosis of common chromosome aneuploidies by QF-PCR. Assessment on 18,000 consecutive clinical samples. Mol Hum Reprod. 2004; 10:839–846.15. Donaghue C, Roberts A, Mann K, Ogilvie CM. Development and targeted application of a rapid QF-PCR test for sex chromosome imbalance. Prenat Diagn. 2003; 23:201–210.16. Ledbetter DH, Zachary JM, Simpson JL, Golbus MS, Pergament E, Jackson L, et al. Cytogenetic results from the U.S. Collaborative Study on CVS. Prenat Diagn. 1992; 12:317–345.17. Kalousek DK, Vekemans M. Confined placental mosaicism. J Med Genet. 1996; 33:529–533.18. Stetten G, Escallon CS, South ST, McMichael JL, Saul DO, Blakemore KJ. Reevaluating confined placental mosaicism. Am J Med Genet A. 2004; 131:232–239.19. Donaghue C, Mann K, Docherty Z, Ogilvie CM. Detection of mosaicism for primary trisomies in prenatal samples by QF-PCR and karyotype analysis. Prenat Diagn. 2005; 25:65–72.20. Holgado E, Liddle S, Ballard T, Levett L. Incidence of placental mosaicism leading to discrepant results between QF-PCR and karyotyping in 22,825 chorionic villus samples. Prenat Diagn. 2011; 31:1029–1038.21. Hahnemann JM, Vejerslev LO. Accuracy of cytogenetic findings on chorionic villus sampling (CVS): diagnostic consequences of CVS mosaicism and non-mosaic discrepancy in centres contributing to EUCROMIC 1986-1992. Prenat Diagn. 1997; 17:801–820.22. Hills A, Donaghue C, Waters J, Waters K, Sullivan C, Kulkarni A, et al. QF-PCR as a stand-alone test for prenatal samples: the first 2 years' experience in the London region. Prenat Diagn. 2010; 30:509–517.23. Covone AE, Voglino G, Ravazzolo R. Sex chromosome rearrangements leading to partial aneuploidies and mosaicisms: use of QF-PCR for detection and quantification of the involved cell lines. Int J Mol Med. 2004; 14:743–746.24. Soler A, Morales C, Badenas C, Rodriguez-Revenga L, Carrio A, Margarit E, et al. A retrospective and theoretical evaluation of rapid methods for detecting chromosome abnormalities and their implications on genetic counseling based on a series of 3868 CVS diagnoses. Fetal Diagn Ther. 2008; 23:126–131.25. Papp Z. Amniocentesis vs chorionic villous sampling as a diagnostic test after an abnormal noninvasive prenatal testing result. Am J Obstet Gynecol. 2015; 213:881–882.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Obstetrical Outcomes of Amniocentesis or Chorionic Villus Sampling in Dichorionic Twin Pregnancies
- DNA - based Prenatal Diagnosis of Epidermolytic Palmoplantar Keratoderma
- Chorionic villus sampling
- A study of chorionic villus sampling for prenatal diagnosis of chromosomal abnormalities
- Prenatal diagnosis of a fetus with recurrent translocation 21 trisomy by chorionic villus sampling