Yonsei Med J.
2017 Jan;58(1):165-173. 10.3349/ymj.2017.58.1.165.
Certain Polymorphisms in SP110 Gene Confer Susceptibility to Tuberculosis: A Comprehensive Review and Updated Meta-Analysis
- Affiliations
-
- 1Center for Gene Diagnosis, Zhongnan Hospital of Wuhan University, Wuhan, Hubei, China. zhengfang@whu.edu.cn
- 2Hubei Provincial Key Laboratory of Developmentally Originated Disease, Wuhan, Hubei, China.
- KMID: 2374203
- DOI: http://doi.org/10.3349/ymj.2017.58.1.165
Abstract
- PURPOSE
Numerous studies have assessed the association of SP110 gene variants with tuberculosis (TB), but the results were inconsistent. Through a comprehensive review and meta-analysis, our study aimed to clarify the nature of genetic risks contributed by 11 polymorphisms for the development of TB.
MATERIALS AND METHODS
Through searching PubMed, web of science, China National Knowledge Infrastructure (CNKI) databases, a total of 11 articles including 13 independent studies were selected. The pooled odd ratios (ORs) along with their corresponding 95% confidence interval (CI) were estimated for allelic comparisons, additive model (homozygote comparisons; heterozygote comparisons), dominant model and recessive model. We also assessed the heterogeneity across the studies and publication bias.
RESULTS
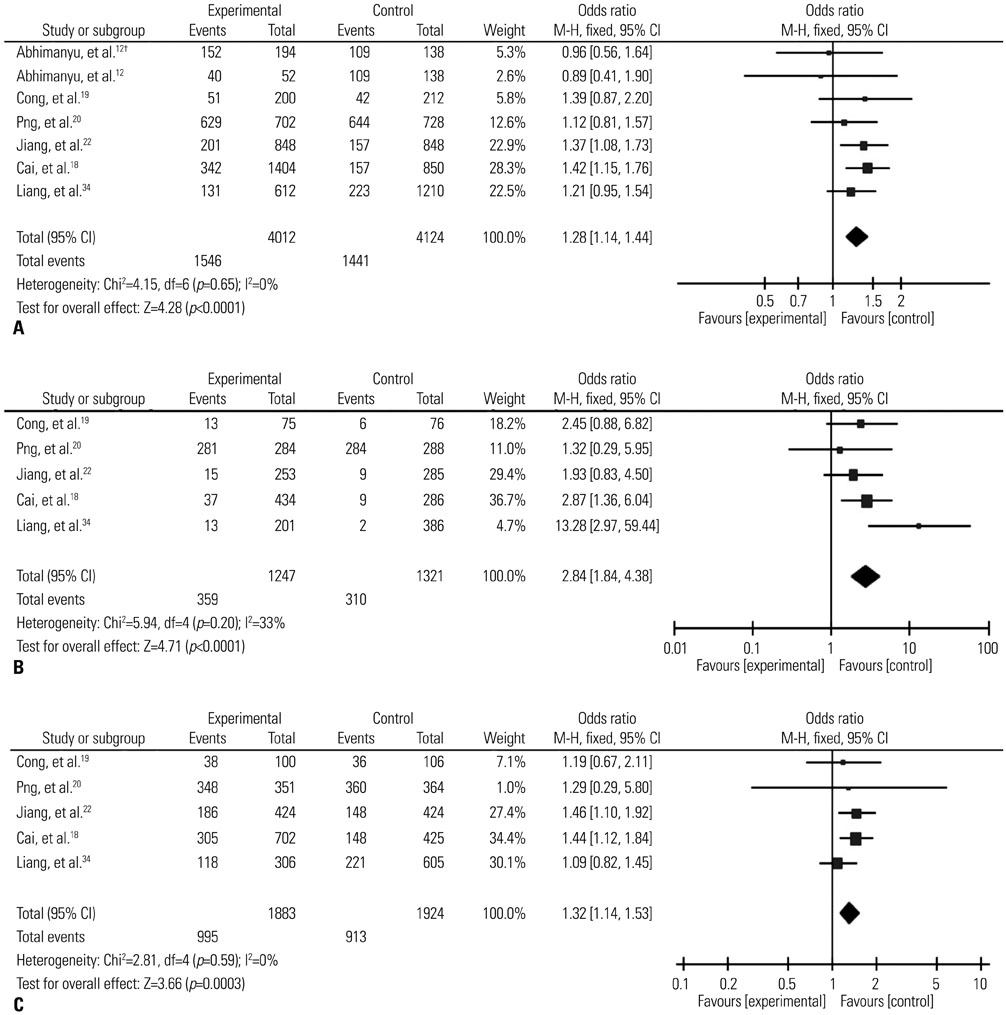
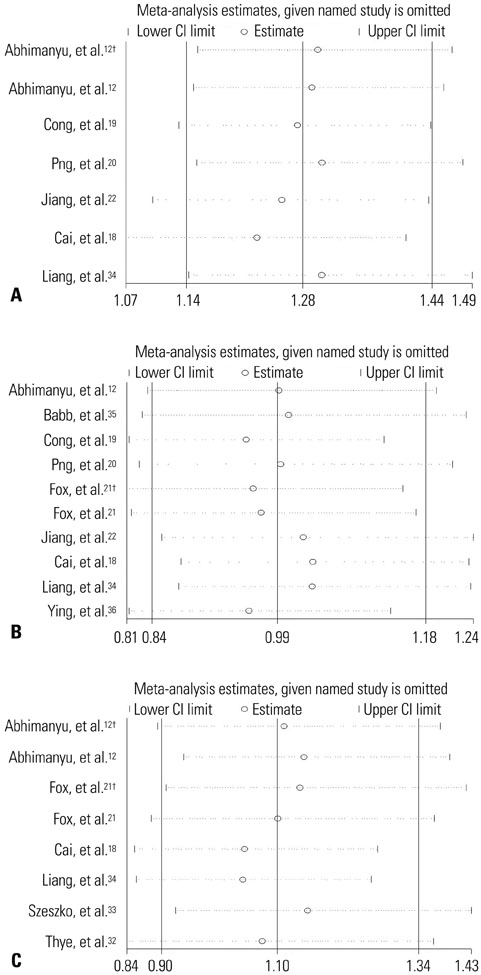
The results of combined analysis revealed a significantly increased risk of TB for single nucleotide polymorphism (SNP) rs9061 in all five comparisons (allelic comparisons: OR=1.28, 95% CI=1.14-1.44, p<0.0001; homozygote comparisons: OR=2.84, 95% CI=1.84-4.38, p<0.00001; heterozygote comparisons: OR=1.23, 95% CI=1.05-1.43, p=0.009; dominant model: OR=1.32, 95% CI=1.14-1.53, p=0.0003; recessive model: OR=2.26, 95% CI=1.18-4.34, p=0.01). In subgroup analysis, the risk of TB associated with SNP rs9061 appeared to be increased. Moreover, increased risk of TB was also found in Asian subgroup of SNP rs11556887, while decreased risk of TB appeared in large sample size subgroup of SNP rs1135791. No significant association was observed between other SNPs and the risk of TB.
CONCLUSION
Our meta-analysis suggested that the variant of SNP rs9061 might be a risk factor for TB.
Keyword
MeSH Terms
-
Alleles
Asian Continental Ancestry Group/genetics
China
Confidence Intervals
Genetic Predisposition to Disease
Heterozygote
Homozygote
Humans
Minor Histocompatibility Antigens/*genetics
Nuclear Proteins/*genetics
Odds Ratio
*Polymorphism, Single Nucleotide
Risk Factors
Tuberculosis, Pulmonary/*genetics
Minor Histocompatibility Antigens
Nuclear Proteins
Figure
Reference
-
1. Sakamoto K. The pathology of Mycobacterium tuberculosis infection. Vet Pathol. 2012; 49:423–439.
Article2. Dye C, Scheele S, Dolin P, Pathania V, Raviglione MC. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA. 1999; 282:677–686.3. Hill AV. Aspects of genetic susceptibility to human infectious diseases. Annu Rev Genet. 2006; 40:469–486.
Article4. Comstock GW. Tuberculosis in twins: a re-analysis of the Prophit survey. Am Rev Respir Dis. 1978; 117:621–624.5. Stead WW, Senner JW, Reddick WT, Lofgren JP. Racial differences in susceptibility to infection by Mycobacterium tuberculosis. N Engl J Med. 1990; 322:422–427.
Article6. Tosh K, Campbell SJ, Fielding K, Sillah J, Bah B, Gustafson P, et al. Variants in the SP110 gene are associated with genetic susceptibility to tuberculosis in West Africa. Proc Natl Acad Sci U S A. 2006; 103:10364–10368.
Article7. Möller M, Hoal EG. Current findings, challenges and novel approaches in human genetic susceptibility to tuberculosis. Tuberculosis (Edinb). 2010; 90:71–83.
Article8. Pan H, Yan BS, Rojas M, Shebzukhov YV, Zhou H, Kobzik L, et al. Ipr1 gene mediates innate immunity to tuberculosis. Nature. 2005; 434:767–772.
Article9. Li N, Liu P, Wang L, Liu J, Yuan X, Meng W, et al. Effect of Ipr1 on expression levels of immune genes related to macrophage anti-infection of mycobacterium tuberculosis. Int J Clin Exp Med. 2015; 8:3411–3419.10. Kramnik I. Genetic dissection of host resistance to Mycobacterium tuberculosis: the sst1 locus and the Ipr1 gene. Curr Top Microbiol Immunol. 2008; 321:123–148.
Article11. Apt AS. Are mouse models of human mycobacterial diseases relevant? Genetics says: ‘yes!’. Immunology. 2011; 134:109–115.
Article12. Abhimanyu , Jha P, Jain A, Arora K, Bose M. Genetic association study suggests a role for SP110 variants in lymph node tuberculosis but not pulmonary tuberculosis in north Indians. Hum Immunol. 2011; 72:576–580.
Article13. Wu H, Wang Y, Zhang Y, Yang M, Lv J, Liu J, et al. TALE nickase-mediated SP110 knockin endows cattle with increased resistance to tuberculosis. Proc Natl Acad Sci U S A. 2015; 112:E1530–E1539.14. Zhou D, Li G. Nuclear body Sp110 and its biological functions. J Med Mol Biol. 2006; 4:271–274.15. Bloch DB, Nakajima A, Gulick T, Chiche JD, Orth D, de La Monte SM, et al. Sp110 localizes to the PML-Sp100 nuclear body and may function as a nuclear hormone receptor transcriptional coactivator. Mol Cell Biol. 2000; 20:6138–6146.
Article16. Castrillo A, Tontonoz P. Nuclear receptors in macrophage biology: at the crossroads of lipid metabolism and inflammation. Annu Rev Cell Dev Biol. 2004; 20:455–480.
Article17. Bellamy R. Genetic susceptibility to tuberculosis. Clin Chest Med. 2005; 26:233–246.
Article18. Cai L, Deng SL, Liang L, Pan H, Zhou J, Wang MY, et al. Identification of genetic associations of SP110/MYBBP1A/RELA with pulmonary tuberculosis in the Chinese Han population. Hum Genet. 2013; 132:265–273.
Article19. Cong J, Li G, Zhou D, Tao Y, Xiong Y. [Study on relation between Sp110 gene polymorphism and tuberculosis genetic susceptibility of Chongqing Han People]. Wei Sheng Yan Jiu. 2010; 39:540–544.20. Png E, Alisjahbana B, Sahiratmadja E, Marzuki S, Nelwan R, Adnan I, et al. Polymorphisms in SP110 are not associated with pulmonary tuberculosis in Indonesians. Infect Genet Evol. 2012; 12:1319–1323.
Article21. Fox GJ, Sy DN, Nhung NV, Yu B, Ellis MK, Van Hung N, et al. Polymorphisms of SP110 are associated with both pulmonary and extrapulmonary tuberculosis among the Vietnamese. PLoS One. 2014; 9:e99496.
Article22. Jiang SY, Li LL, Yue J, Chen WZ, Yang C, Wan CL, et al. The effects of SP110’s associated genes on fresh cavitary pulmonary tuberculosis in Han Chinese population. Clin Exp Med. 2016; 16:219–225.
Article23. Persson C, Canedo P, Machado JC, El-Omar EM, Forman D. Polymorphisms in inflammatory response genes and their association with gastric cancer: a HuGE systematic review and meta-analyses. Am J Epidemiol. 2011; 173:259–270.
Article24. Lu XC, Yu W, Tao Y, Zhao PL, Li K, Tang LJ, et al. Contribution of transforming growth factor α polymorphisms to nonsyndromic orofacial clefts: a HuGE review and meta-analysis. Am J Epidemiol. 2014; 179:267–281.
Article25. Liang J, Lin C, Hu F, Wang F, Zhu L, Yao X, et al. APC polymorphisms and the risk of colorectal neoplasia: a HuGE review and meta-analysis. Am J Epidemiol. 2013; 177:1169–1179.
Article26. Wu J, Liu J, Zhou Y, Ying J, Zou H, Guo S, et al. Predictive value of XRCC1 gene polymorphisms on platinum-based chemotherapy in advanced non-small cell lung cancer patients: a systematic review and meta-analysis. Clin Cancer Res. 2012; 18:3972–3981.
Article27. Thakkinstian A, McEvoy M, Minelli C, Gibson P, Hancox B, Duffy D, et al. Systematic review and meta-analysis of the association between {beta}2-adrenoceptor polymorphisms and asthma: a HuGE review. Am J Epidemiol. 2005; 162:201–211.
Article28. DerSimonian R, Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials. 2007; 28:105–114.
Article29. Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997; 127:820–826.
Article30. Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994; 50:1088–1101.
Article31. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997; 315:629–634.
Article32. Thye T, Browne EN, Chinbuah MA, Gyapong J, Osei I, Owusu-Dabo E, et al. No associations of human pulmonary tuberculosis with Sp110 variants. J Med Genet. 2006; 43:e32.33. Szeszko JS, Healy B, Stevens H, Balabanova Y, Drobniewski F, Todd JA, et al. Resequencing and association analysis of the SP110 gene in adult pulmonary tuberculosis. Hum Genet. 2007; 121:155–160.
Article34. Liang L, Zhao YL, Yue J, Liu JF, Han M, Wang H, et al. Association of SP110 gene polymorphisms with susceptibility to tuberculosis in a Chinese population. Infect Genet Evol. 2011; 11:934–939.
Article35. Babb C, Keet EH, van Helden PD, Hoal EG. SP110 polymorphisms are not associated with pulmonary tuberculosis in a South African population. Hum Genet. 2007; 121:521–522.
Article36. Ying X, Hui L, Yu HD, Jie L, Jing S, Long W, et al. Interaction of SP110 and VDR gene polymorphisms with environmental factors in tuberculosis. J Reg Anat Oper Sorg. 2013; 04:377–379.37. Russell DG, Barry CE 3rd, Flynn JL. Tuberculosis: what we don’t know can, and does, hurt us. Science. 2010; 328:852–856.
Article38. Lei X, Zhu H, Zha L, Wang Y. SP110 gene polymorphisms and tuberculosis susceptibility: a systematic review and meta-analysis based on 10 624 subjects. Infect Genet Evol. 2012; 12:1473–1480.
Article39. Deléage G, Combet C, Blanchet C, Geourjon C. ANTHEPROT: an integrated protein sequence analysis software with client/server capabilities. Comput Biol Med. 2001; 31:259–267.
Article40. Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014; 11:361–362.
Article41. Dechartres A, Trinquart L, Boutron I, Ravaud P. Influence of trial sample size on treatment effect estimates: meta-epidemiological study. BMJ. 2013; 346:f2304.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Impact of CYP1A1 variants on the risk of acute lymphoblastic leukemia: evidence from an updated meta‑analysis
- Circulating VEGF levels and genetic polymorphisms in Behçet’s disease: a meta-analysis
- Vitamin D Receptor Gene, Matrix Metalloproteinase 3 Polymorphisms and the Risk of Intervertebral Disc Degeneration Susceptibility: Meta-Analysis
- IL-10 Polymorphisms and Tuberculosis Susceptibility: An Updated Meta-Analysis
- The Link between Suicidality and Electroencephalography Asymmetry: A Systematic Review and Meta-analysis