J Gastric Cancer.
2010 Sep;10(3):91-98.
Genetic and Expression Analysis of the SIRT1 Gene in Gastric Cancers
- Affiliations
-
- 1Department of Pathology, The Catholic University of Korea, School of Medicine, Seoul, Korea. wonsang@catholic.ac.kr
- 2Department of Pathology, Binzhou Medical College, Binzhou, China.
Abstract
- PURPOSE
Silent mating-type information regulation 2 homologue 1 (SIRT1) is a nicotinamide adenine dinucleotide-dependent deacetylase. SIRT1 plays an important role in the regulation of cell death/survival and stress response in mammals. The aim of this study was to investigate whether the SIRT1 gene is involved in the development or progression of gastric cancers.
MATERIALS AND METHODS
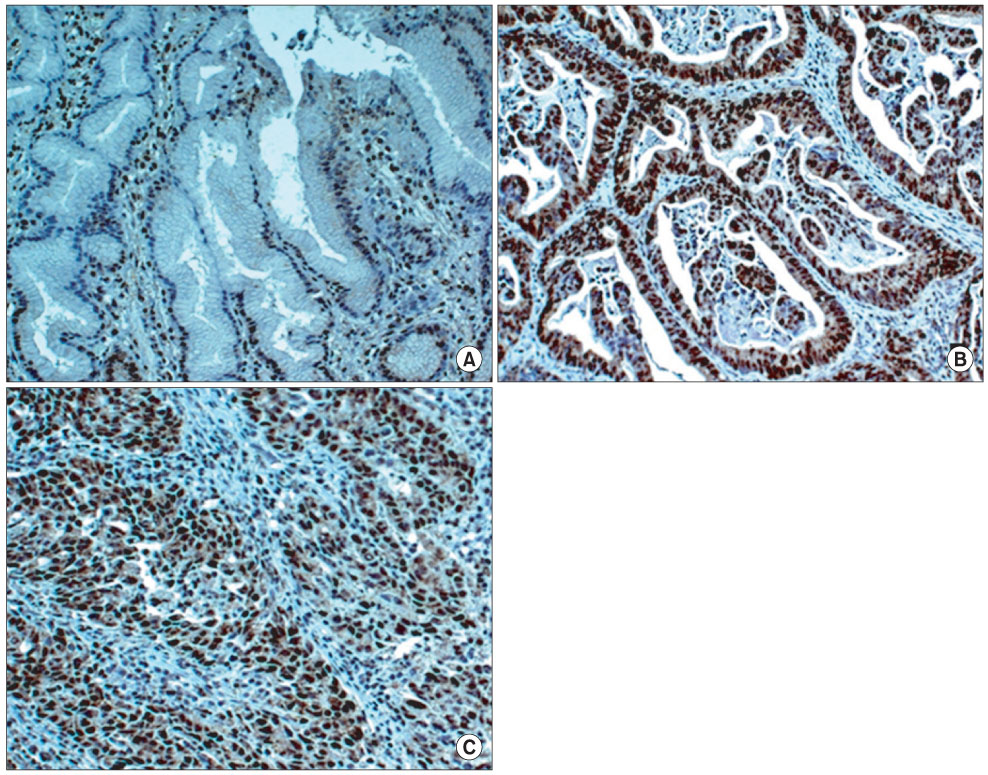
SIRT1 and p53 genes in 86 gastric cancers were examined for genetic alterations by PCR-single strand conformation polymorphism sequencing, as well as SIRT1 protein expression in 170 gastric cancers by immunohistochemistry.
RESULTS
In the genetic analysis, we found SIRT1 and p53 mutations in two and 12 cases, respectively. Two missense mutations, c.599 C>T (T200I) and c.1258 G>A (E420K), were detected in the SIRT1 gene coding region. The SIRT1 and p53 mutation were found in mutually exclusive gastric cancers. The immunohistochemistry revealed that SIRT1 overexpression was found in 95 (55.9%) of 170 gastric cancers. Altered SIRT1 expression was not statistically associated with clinicopathological parameters, including tumor differentiation, location, lymph node metastasis, or p53 expression. Two cases with an SIRT1 mutation showed increased SIRT1 expression.
CONCLUSIONS
These results suggest that genetic alterations and overexpression of the SIRT1 gene may contribute to gastric cancer development.
Keyword
MeSH Terms
Figure
Reference
-
1. Shin HR, Jung KW, Won YJ, Park JG. 139 KCCR-affiliated Hospitals. 2002 annual report of the Korea Central Cancer Registry: based on registered data from 139 hospitals. Cancer Res Treat. 2004. 36:103–114.
Article2. Yuasa Y. Control of gut differentiation and intestinal-type gastric carcinogenesis. Nat Rev Cancer. 2003. 3:592–600.
Article3. Bordone L, Cohen D, Robinson A, Motta MC, van Veen E, Czopik A, et al. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007. 6:759–767.
Article4. Murayama A, Ohmori K, Fujimura A, Minami H, Yasuzawa-Tanaka K, Kuroda T, et al. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008. 133:627–639.
Article5. Hasegawa K, Yoshikawa K. Necdin regulates p53 acetylation via Sirtuin1 to modulate DNA damage response in cortical neurons. J Neurosci. 2008. 28:8772–8784.
Article6. Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007. 23:413–418.
Article7. Fraga MF, Agrelo R, Esteller M. Cross-talk between aging and cancer: the epigenetic language. Ann N Y Acad Sci. 2007. 1100:60–74.
Article8. Lim CS. Human SIRT1: a potential biomarker for tumorigenesis? Cell Biol Int. 2007. 31:636–637.
Article9. Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008. 135:907–918.
Article10. Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008. 14:312–323.
Article11. Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, et al. hSIR2 (SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001. 107:149–159.
Article12. Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004. 303:2011–2015.
Article13. Lee JH, Song MY, Song EK, Kim EK, Moon WS, Han MK, et al. Overexpression of SIRT1 protects pancreatic beta-cells against cytokine toxicity by suppressing the nuclear factor-kappaB signaling pathway. Diabetes. 2009. 58:344–351.
Article14. Cohen HY, Lavu S, Bitterman KJ, Hekking B, Imahiyerobo TA, Miller C, et al. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol Cell. 2004. 13:627–638.
Article15. Langley E, Pearson M, Faretta M, Bauer UM, Frye RA, Minucci S, et al. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002. 21:2383–2396.
Article16. Luo J, Li M, Tang Y, Laszkowska M, Roeder RG, Gu W. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci U S A. 2004. 101:2259–2264.
Article17. Ford J, Jiang M, Milner J. Cancer-specific functions of SIRT1 enable human epithelial cancer cell growth and survival. Cancer Res. 2005. 65:10457–10463.
Article18. Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004. 306:2105–2108.
Article19. Kononen J, Bubendorf L, Kallioniemi A, Bärlund M, Schraml P, Leighton S, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998. 4:844–847.
Article20. Lee JY, Dong SM, Kim SY, Yoo NJ, Lee SH, Park WS. A simple, precise and economical microdissection technique for analysis of genomic DNA from archival tissue sections. Virchows Arch. 1998. 433:305–309.
Article21. Kim CJ, Song JH, Cho YG, Cao Z, Kim SY, Nam SW, et al. Activation-induced cytidine deaminase expression in gastric cancer. Tumour Biol. 2007. 28:333–339.
Article22. Jang KY, Hwang SH, Kwon KS, Kim KR, Choi HN, Lee NR, et al. SIRT1 expression is associated with poor prognosis of diffuse large B-cell lymphoma. Am J Surg Pathol. 2008. 32:1523–1531.
Article23. Ford J, Jiang M, Milner J. Cancer-specific functions of SIRT1 enable human epithelial cancer cell growth and survival. Cancer Res. 2005. 65:10457–10463.
Article24. Ota H, Tokunaga E, Chang K, Hikasa M, Iijima K, Eto M, et al. Sirt1 inhibitor, Sirtinol, induces senescence-like growth arrest with attenuated Ras-MAPK signaling in human cancer cells. Oncogene. 2006. 25:176–185.
Article25. Uchino S, Noguchi M, Ochiai A, Saito T, Kobayashi M, Hirohashi S. p53 mutation in gastric cancer: a genetic model for carcinogenesis is common to gastric and colorectal cancer. Int J Cancer. 1993. 54:759–764.
Article26. Sheffield VC, Beck JS, Kwitek AE, Sandstrom DW, Stone EM. The sensitivity of single-strand conformation polymorphism analysis for the detection of single base substitutions. Genomics. 1993. 16:325–332.
Article27. Williams C, Pontén F, Moberg C, Söderkvist P, Uhlén M, Pontén J, et al. A high frequency of sequence alterations is due to formalin fixation of archival specimens. Am J Pathol. 1999. 155:1467–1471.
Article28. Cha EJ, Noh SJ, Kwon KS, Kim CY, Park BH, Park HS, et al. Expression of DBC1 and SIRT1 is associated with poor prognosis of gastric carcinoma. Clin Cancer Res. 2009. 15:4453–4459.
Article29. Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005. 16:4623–4635.
Article30. Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS One. 2008. 3:e2020.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- SIRT1 Inhibits p53 but not NF-kappaB Transcriptional Activity during Differentiation of Mouse Embryonic Stem Cells into Embryoid Bodies
- Expression of SIRT1 and DBC1 in Gastric Adenocarcinoma
- Effect of Resveratrol, a SIRT1 Activator, on the Interactions of the CLOCK/BMAL1 Complex
- Prognostic Significance of Sirtuins Expression in Papillary Thyroid Carcinoma
- Immunohistochemical Study of p53 and nm23-H1 Protein in Gastric Carcinoma