Ann Lab Med.
2015 Jan;35(1):132-136. 10.3343/alm.2015.35.1.132.
Clinical Relevance of High-Resolution Single Nucleotide Polymorphism Array in Patients with Relapsed Acute Lymphoblastic Leukemia with Normal Karyotype: A Report of Three Cases
- Affiliations
-
- 1Department of Laboratory Medicine, Pusan National University School of Medicine, Pusan National University Hospital, Busan, Korea. eylee@pusan.ac.kr
- 2Biomedical Research Institute, Pusan National University Hospital, Busan, Korea.
- 3Department of Laboratory Medicine, Pusan National University School of Medicine, Pusan National University Yangsan Hospital, Yangsan, Korea.
- KMID: 2363161
- DOI: http://doi.org/10.3343/alm.2015.35.1.132
Abstract
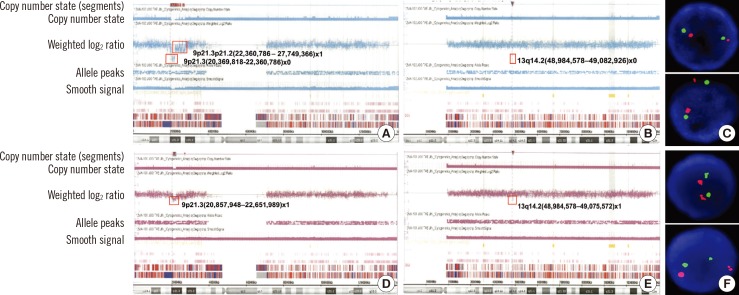
- We report three patients with normal karyotype (NK) ALL, who showed genetic aberrations as determined by high-resolution single nucleotide polymorphism array (SNP-A) analysis at both diagnosis and relapse. We evaluated the clinical relevance of the SNP-A assay for the detection of subtle changes in the size of affected genetic lesions at relapse as well as the prognostic value of the assay. In our patients, application of the SNP-A assay enabled sensitive detection of cryptic changes affecting clinically important genes in NK ALL. Therefore, this assay seems to be more advantageous compared to other conventional methods such as FISH assay, HemaVision (DNA Technology, Denmark), and conventional karyotyping for the detection of an "unstable genotype" at relapse, which may be associated with microscopic clonal evolution and poor prognosis. Further comprehensive studies are required to confirm the issues presented by our case patients in this report.
Keyword
MeSH Terms
-
Adult
Cyclin-Dependent Kinase Inhibitor p16/genetics
Female
Genotype
Humans
In Situ Hybridization, Fluorescence
Karyotype
Karyotyping
Loss of Heterozygosity
Male
Middle Aged
Oligonucleotide Array Sequence Analysis
Polymorphism, Single Nucleotide
Precursor Cell Lymphoblastic Leukemia-Lymphoma/*diagnosis/genetics
Recurrence
Retinoblastoma Protein/genetics
Cyclin-Dependent Kinase Inhibitor p16
Retinoblastoma Protein
Figure
Reference
-
1. Bullinger L, Krönke J, Schön C, Radtke I, Urlbauer K, Botzenhardt U, et al. Identification of acquired copy number alterations and uniparental disomies in cytogenetically normal acute myeloid leukemia using high-resolution single-nucleotide polymorphism analysis. Leukemia. 2010; 24:438–449. PMID: 20016533.
Article2. Barresi V, Romano A, Musso N, Capizzi C, Consoli C, Martelli MP, et al. Broad copy neutral-loss of heterozygosity regions and rare recurring copy number abnormalities in normal karyotype-acute myeloid leukemia genomes. Genes Chromosomes Cancer. 2010; 49:1014–1023. PMID: 20725993.
Article3. Tiu RV, Gondek LP, O'Keefe CL, Huh J, Sekeres MA, Elson P, et al. New lesions detected by single nucleotide polymorphism array-based chromosomal analysis have important clinical impact in acute myeloid leukemia. J Clin Oncol. 2009; 27:5219–5226. PMID: 19770377.
Article4. Yi JH, Huh J, Kim HJ, Kim SH, Kim HJ, Kim YK, et al. Adverse prognostic impact of abnormal lesions detected by genome-wide single nucleotide polymorphism array-based karyotyping analysis in acute myeloid leukemia with normal karyotype. J Clin Oncol. 2011; 29:4702–4708. PMID: 22084373.
Article5. Hahm C, Huh HJ, Mun YC, Seong CM, Chung WS, Huh J. Genomic aberrations of myeloproliferative and myelodysplastic/myeloproliferative neoplasms in chronic phase and during disease progression. Int J Lab Hematol. 2014; 5. 21. doi: 10.1111/ijlh.12257. [Epub ahead of print].
Article6. Huh J, Jung CW, Kim HJ, Kim YK, Moon JH, Sohn SK, et al. Different characteristics identified by single nucleotide polymorphism array analysis in leukemia suggest the need for different application strategies depending on disease category. Genes Chromosomes Cancer. 2013; 52:44–55. PMID: 23023762.
Article7. Betts D. del(13q) in ALL. Atlas Genet Cytogenet Oncol Haematol. 2005; 9:24–25.
Article8. Kim M, Yim SH, Cho NS, Kang SH, Ko DH, Oh B, et al. Homozygous deletion of CDKN2A (p16, p14) and CDKN2B (p15) genes is a poor prognostic factor in adult but not in childhood B-lineage acute lymphoblastic leukemia: a comparative deletion and hypermethylation study. Cancer Genet Cytogenet. 2009; 195:59–65. PMID: 19837270.
Article9. Heerema NA. 9p rearrangements in ALL. Atlas Genet Cytogenet Oncol Haematol. 1999; 3:153–154. (http://atlasgeneticsoncology.org/Anomalies/9prearrALLID1156.html).
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Ring Chromosome 5 in Acute Myeloid Leukemia Defined by Whole-genome Single Nucleotide Polymorphism Array
- A Case of Lineage Switch from Acute Lymphoblastic Leukemia to Acute Myeloid Leukemia
- Precursor B-Cell Acute Lymphoblastic Leukemia in Two Patients with a History of Cytotoxic Therapy
- Recent advances in the treatment of adults with acute lymphoblastic leukemia
- Clinical Outcome of the Chromosomal Abnormalities in Adult Acute Lymphoblastic Leukemia (Preliminary Data)