Ann Clin Microbiol.
2016 Dec;19(4):110-120. 10.5145/ACM.2016.19.4.110.
Taxonomic Identification of Bacillus Species Using Matrix-Assisted Laser Desorption/Ionization-Time of Flight Mass Spectrometry
- Affiliations
-
- 1Pathogen Resource TF, Center for Infectious Diseases, Korea National Institute of Health, Korea Centers for Disease Control and Prevention, Cheongju, Korea. kyuhwang61@korea.kr
- KMID: 2362957
- DOI: http://doi.org/10.5145/ACM.2016.19.4.110
Abstract
- BACKGROUND
In this study, we compared various methods of taxonomic identification of Bacillus strains: biochemical methods, 16S rRNA gene sequencing, and matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS). We also developed a pathogen- isolate resource database, thus increasing the identification rate when using MALDI-TOF MS.
METHODS
Thirty Bacillus strains were obtained from the NCCP (National Culture Collection for Pathogens) and were identified using the VITEK 2 system (bio-Mérieux, France), API kit (bioMérieux, France), 16S rRNA gene sequencing, and MALDI-TOF MS. The pathogenicity of Bacillus cereus was confirmed through the identification of virulent genes using a multiplex PCR, and both protein extraction for protein profiling in MALDI-TOF MS and repetitive-sequence fingerprinting were performed.
RESULTS
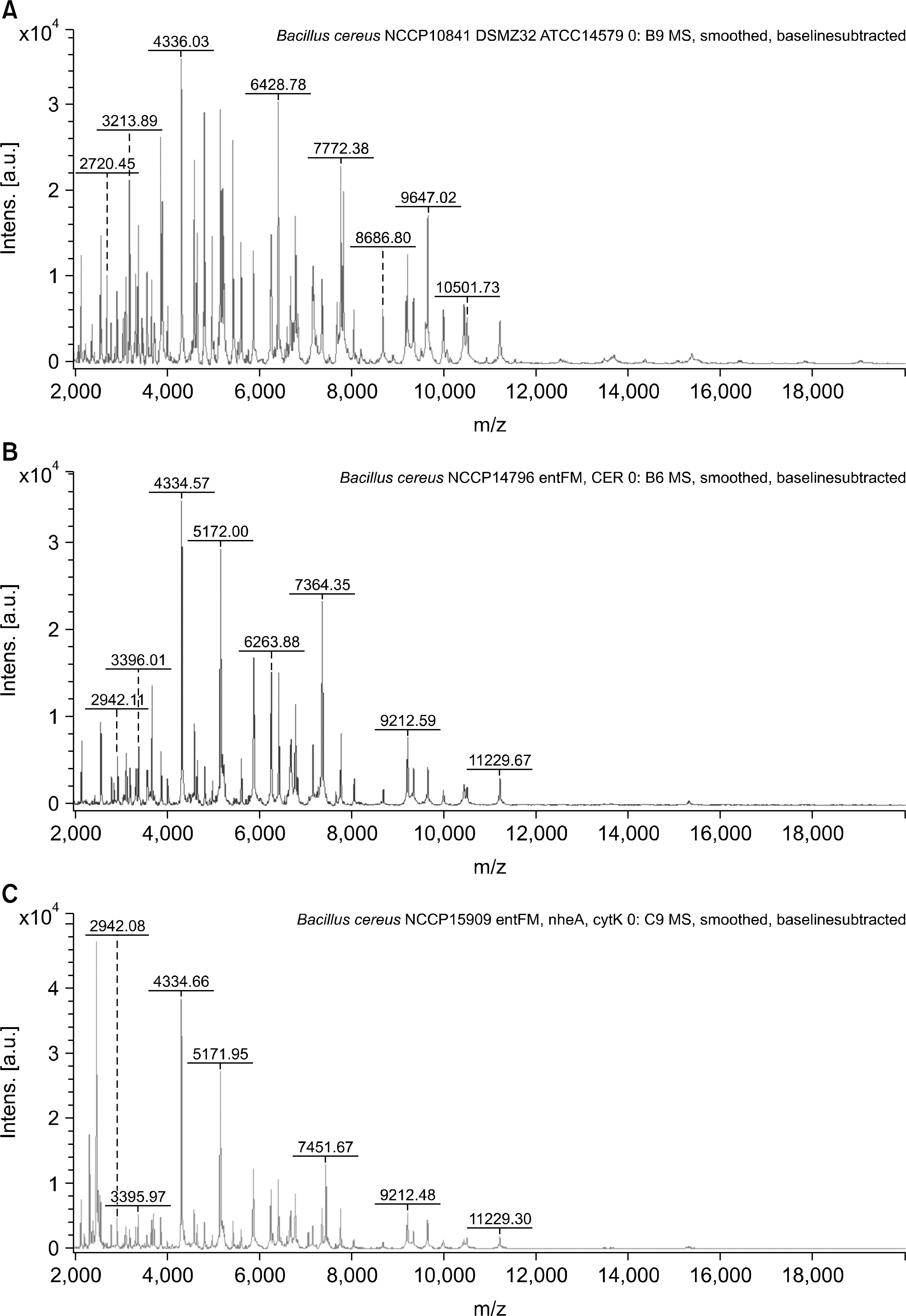
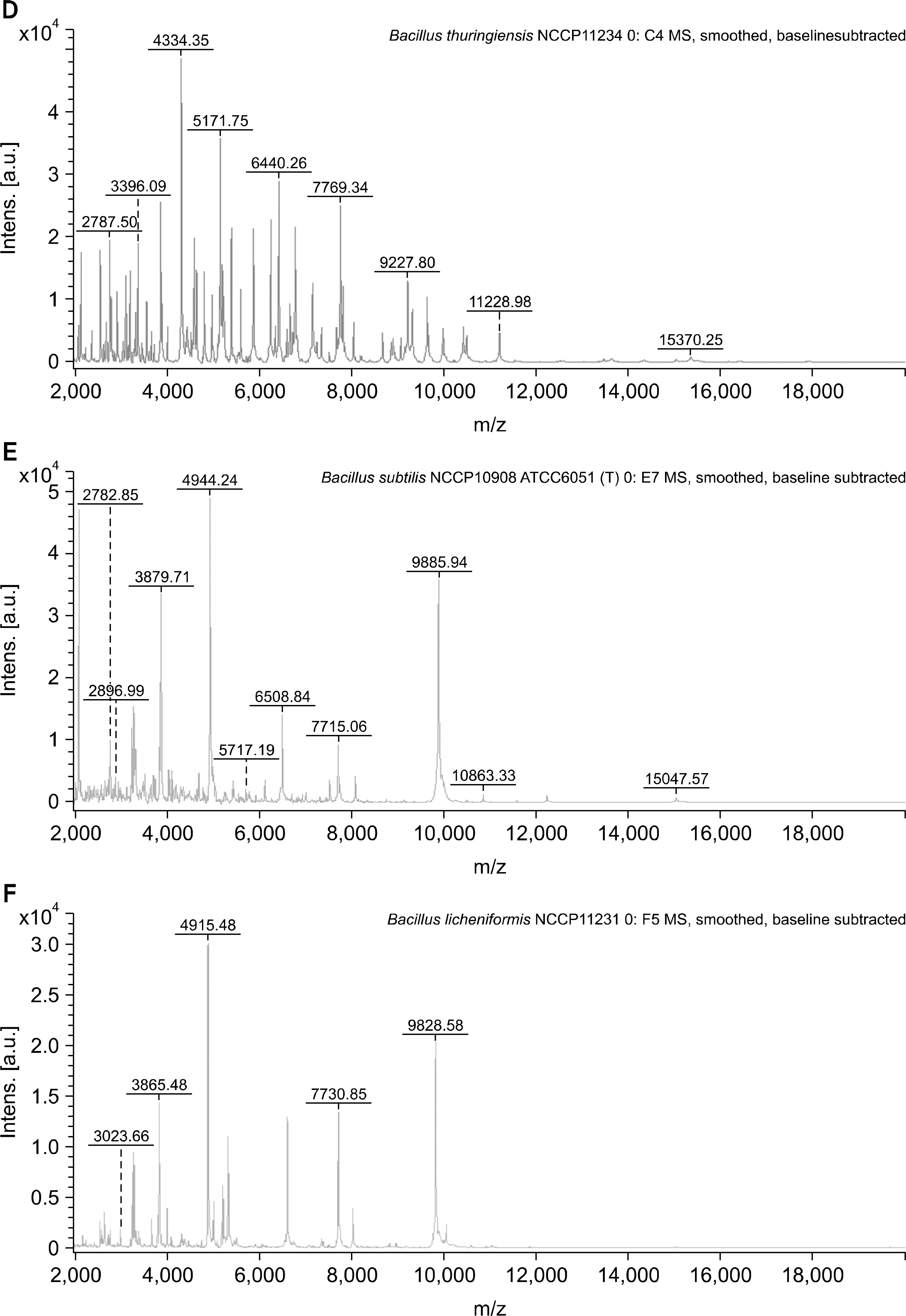
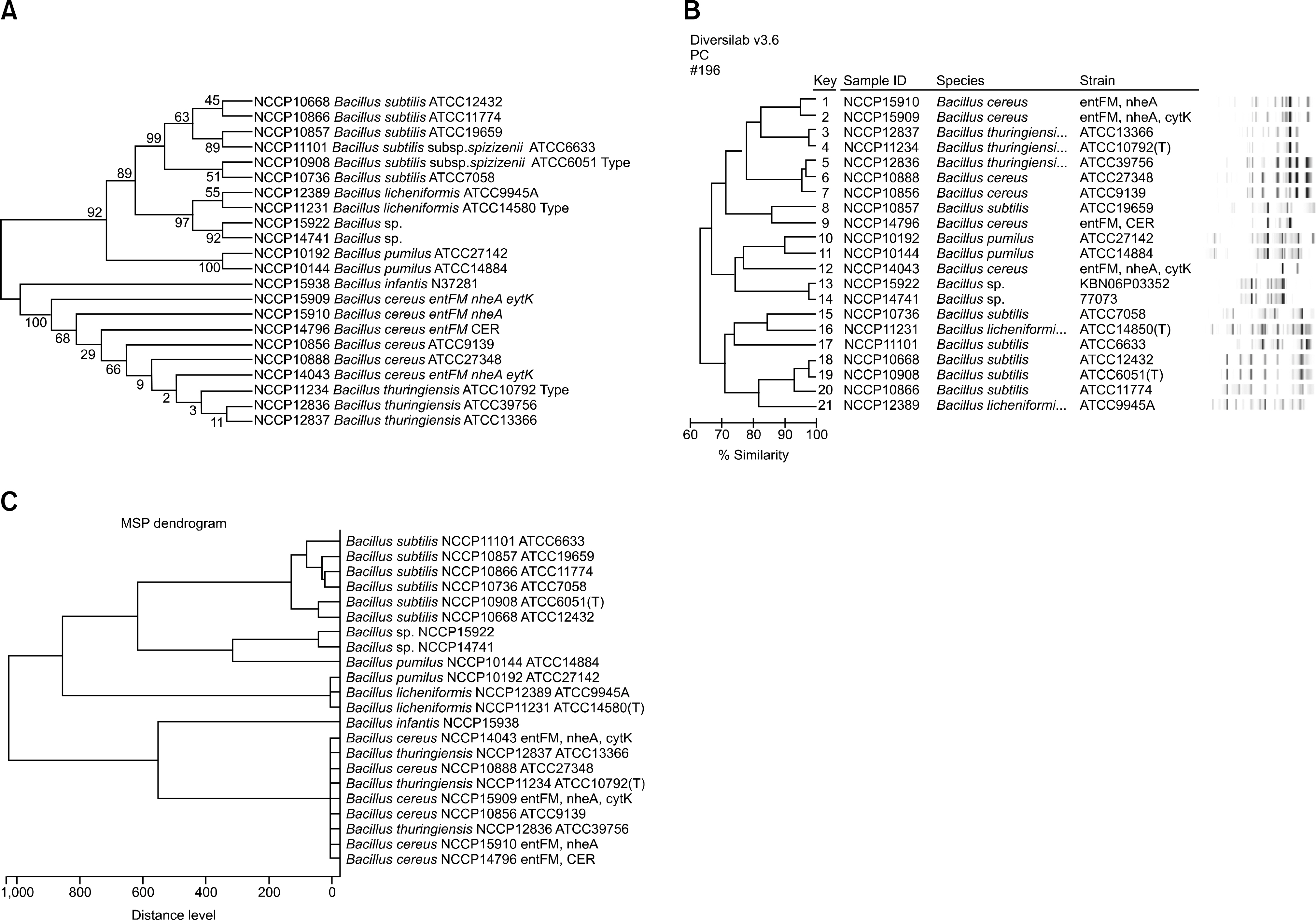
The identification rates at the species level were 40%, 80%, and 76.3% for the VITEK 2 system (bioMérieux), 16S rRNA gene sequencing, and MALDI-TOF MS, respectively. When the major spectrum-profiling dendrogram was compared with the phylogenetic tree, which was constructed based on the 16S rRNA gene sequences and rep-PCR fingerprinting, the classifications were confirmed to be effective.
CONCLUSION
Identification of Bacillus strains using MALDI-TOF MS was more effective than that using the VITEK 2 system (bioMérieux), but was similar to that using 16S rRNA gene sequencing. Continual addition to a proteome-based database can result in increased identification rates for MALDI-TOF MS.
Keyword
MeSH Terms
Figure
Reference
-
References
1. Oguntoyinbo FA. Monitoring of marine bacillus diversity among the bacteria community of sea water. Afr J Biotechnol. 2007; 6:163–6.2. Durak MZ, Fromm HI, Huck JR, Zadoks RN, Boor KJ. Development of molecular typing methods for Bacillus spp. and Paenibacillus spp. isolated from fluid milk products. J Food Sci. 2006; 71:M50–6.
Article3. Fritze D. Taxonomy of the genus bacillus and related genera: the aerobic endospore-forming bacteria. Phytopathology. 2004; 94:1245–8.
Article4. Bavykin SG, Lysov YP, Zakhariev V, Kelly JJ, Jackman J, Stahl DA, et al. Use of 16S rRNA, 23S rRNA, and gyrB gene sequence analysis to determine phylogenetic relationships of Bacillus cereus group microorganisms. J Clin Microbiol. 2004; 42:3711–30.5. Granum PE. Bacillus cereus. Doyle MP, editor. Food Microbiology: Fundamental and Frontiers. 2nd ed.Washington DC: ASM Press;2001. p. 373–81.
Article6. Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, et al. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998; 62:775–806.7. Salkinoja-Salonen M, Vuorio R, Andersson M, Kampfer P, Honkanen-Buzalski T. Toxigenic strains of Bacillus licheniformis related to food poisoning. Appl Environ Microbiol. 1999; 65:4637–45.8. Sorokulova IB, Reva ON, Smirnov VV, Pinchuk IV, Lapa SV, Urdaci MC. Genetic diversity and involvement in bread spoilage of Bacillus strains isolated from flour and ropy bread. Lett Appl Microbiol. 2003; 37:169–73.
Article9. Pavic S, Brett M, Petric I, Lastre D, Smoljanovic M, Atkinson M, et al. An outbreak of food poisoning in a kindergarten caused by milk powder containing toxigenic Bacillus subtilis and Bacillus licheniformis. Archiv für Lebensmittelhygiene. 2005; 56:20–2.10. Jamal W, Albert MJ, Rotimi VO. Realtime comparative evaluation of bioMerieux VITEK MS versus Bruker Microflex MS, two matrix-assisted laser desorption-ionization time-of-flight mass spectrometry systems, for identification of clinically significant bacteria. BMC Microbiol. 2014; 14:289.
Article11. CLSI. Interpretative criteria for identification of bacteria and fungi by DNA target sequencing; approved guideline. CLSI document MM18-A. Pennsylvania: Clinical and Laboratory Standards Institute;2008. p. 1–71.12. Christner M, Trusch M, Rohde H, Kwiatkowski M, Schlüter H, Wolters M, et al. Rapid MALDI-TOF mass spectrometry strain typing during a large outbreak of Shiga-toxigenic Escherichia coli. PLoS One. 2014; 9:e101924.
Article13. Josten M, Reif M, Szekat C, Al-Sabti N, Roemer T, Sparbier K, et al. Analysis of the matrix-assisted laser desorption ionization-time of flight mass spectrum of Staphylococcus aureus identifies mutations that allow differentiation of the main clonal lineages. J Clin Microbiol. 2013; 51:1809–17.
Article14. Khot PD and Fisher MA. Novel approach for differentiating Shigella species and Escherichia coli by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2013; 51:3711–6.15. Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, et al. Introducing EzTaxone-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012; 62:716–21.16. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013; 30:2725–9.
Article17. Kumar S, Tamura K, Nei M. MEGA3: intergrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004; 5:150–63.18. Jukes TH and Cantor CR. Evolution of Protein Molecules. Munro HN, editor. Mammalian Protein Metabolism. New York: Academic Press;1969. p. 21–132.19. Saitou N and Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987; 4:406–25.20. Haigh J, Degun A, Eydmann M, Millar M, Wilks M. Improved performance of bacterium and yeast identification by a commercial matrix-assisted laser desorption ionization-time of flight mass spectrometry system in the clinical microbiology laboratory. J Clin Microbiol. 2011; 49:3441.
Article21. Schulthess B, Bloemberg GV, Zbinden R, Böttger EC, Hombach M. Evaluation of the bruker MALDI biotyper for identification of gram-positive rods: development of a diagnostic algorithm for the clinical laboratory. J Clin Microbiol. 2014; 52:1089–97.
Article22. Healy M, Huong J, Bittner T, Lising M, Frye S, Raza S, et al. Microbial DNA typing by automated repetitive-sequence-based PCR. J Clin Microbiol. 2005; 4:199–207.
Article23. Stanford TE, Bagley CJ, Solomon PJ. Informed baseline subtraction of proteomic mass spectrometry data aided by a novel sliding window algorithm. Proteome Sci. 2016; 14:19.
Article24. Dupont C, Sivadon-Tardy V, Bille E, Dauphin B, Beretti JL, Alvarez AS, et al. Identification of clinical coagulase-negative staphylococci, isolated in microbiology laboratories, by matrix-assisted laser desorption/ionization-time of flight mass spectrometry and two automated systems. Clin Microbiol Infect. 2010; 16:998–1004.
Article25. Benagli C, Demarta A, Caminada A, Ziegler D, Petrini O, Tonolla M. A rapid MALDI-TOF MS identification database at geno-species level for clinical and environmental Aeromonas strains. PLoS One. 2012; 7:e48441.
Article26. Lista F, Reubsaet FA, De Santis R, Parchen RR, de Jong AL, Kieboom J, et al. Reliable identification at the species level of Brucella isolates with MALDI-TOF-MS. BMC Microbiol. 2011; 11:267.
Article27. Lasch P, Beyer W, Nattermann H, Stämmler M, Siegbrecht E, Grunow R, et al. Identification of Bacillus anthracis by using matrix-assisted laser desorption ionization-time of flight mass spectrometry and artificial neural networks. Appl Environ Microbiol. 2009; 75:7229–42.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Reliability of Acinetobacter baumannii Identification with Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry
- Comparison of the Bruker Biotyper and VITEK MS Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry Systems Using a Formic Acid Extraction Method to Identify Common and Uncommon Yeast Isolates
- MALDI-TOF-MS Fingerprinting Provides Evidence of Urosepsis caused by Aerococcus urinae
- Evaluation of Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry for Identification of Aerobic Bacteria in a Clinical Microbiology Laboratory
- Application of Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Imaging Mass Spectrometry (MALDI-TOF IMS) for Premalignant Gastrointestinal Lesions