J Korean Assoc Oral Maxillofac Surg.
2010 Dec;36(6):460-465.
Mesenchymal Smad4 mediated signaling is essential for palate development
- Affiliations
-
- 1Department of Oral and Maxillofacial Surgery, Institute of Oral Bioscience and BK21 program, School of Dentistry, Chonbuk National University, Jeonju, Korea. omfskso@jbnu.ac.kr
Abstract
- INTRODUCTION
A cleft palate is a common birth defect in humans with an incidence of 1/500 to 1/1,000 births. It appears to be caused by multiple genetic and environmental factors during palatogenesis. Many molecules are involved in palate formation but the biological mechanisms underlying the normal palate formation and cleft palate are unclear. Accumulating evidence suggests that transforming growth factor beta/bone morphogenetic proteins (TGF-beta/BMP) family members mediate the epithelial-mesenchymal interactions during palate formation. However, their roles in palatal morphogenesis are not completely understood.
MATERIALS AND METHODS
To understand the roles of TGF-beta/BMP signaling in vivo during palatogenesis, mice with a palatal mesenchyme-specific deletion of Smad4, a key intracellular mediator of TGF-beta/BMP signaling, were generated and analyzed using the Osr2Ires-Cre mice.
RESULTS
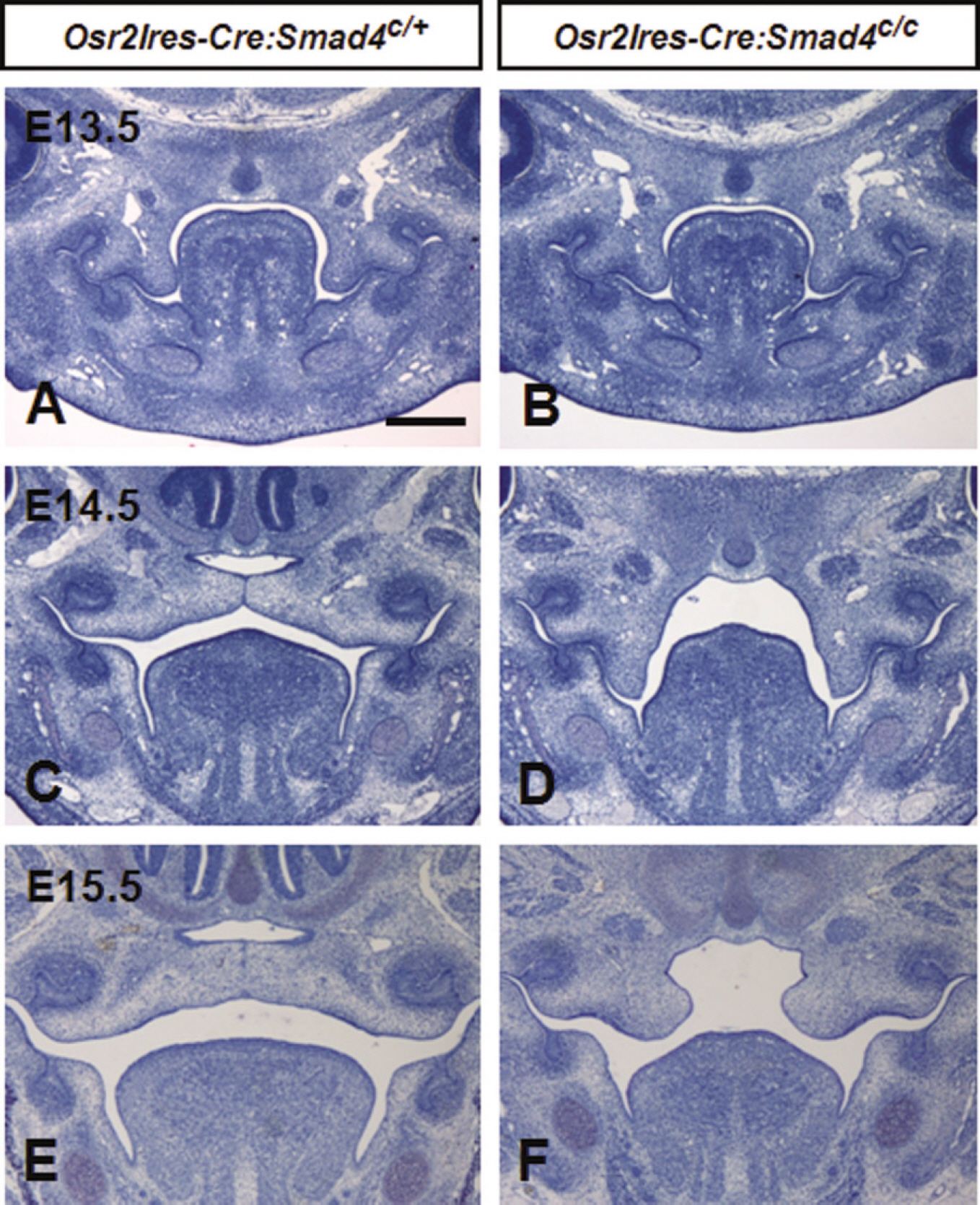
The mutant mice were alive at the time of birth with open eyelids and complete cleft palate but died within 24 hours after birth. In skeletal preparation, the horizontal processes of the palatine bones in mutants were not formed and resulted in a complete cleft palate. At E13.5, the palatal shelves of the mutants were growing as normally as those of theirwild type littermates. However, the palatal shelves of the mutants were not elevated at E14.5 in contrast to the elevated palatal shelves of the wild type mice. At E15.5, the palatal shelves of the mutants were elevated over the tongue but did not come in contact with each other, resulting in a cleft palate.
CONCLUSION
These results suggest that mesenchymal Smad4 mediated signaling is essential for the growth of palatal processes and suggests that TGF-beta/BMP family members are essential regulators during palate development.
Keyword
MeSH Terms
Figure
Reference
-
1. Marazita ML, Field LL, Cooper ME, Tobias R, Maher BS, Peanchitlertkajorn S, et al. Genome scan for loci involved in cleft lip with or without cleft palate, in Chinese multiplex families. Am J Hum Genet. 2002; 71:349–64.
Article2. Gritli-Linde A. Molecular control of secondary palate development. Dev Biol. 2007; 301:309–26.
Article3. Massague′J. How cells read TGF-βsignals. Nat Rev Mol Cell Biol. 2000; 1:169–78.4. Weinstein M, Yang X, Deng C. Functions of mammalian Smad genes as revealed by targeted gene disruption in mice. Cytokine Growth Factor Rev. 2000; 11:49–58.
Article5. Metzger D, Chambon P. Site- and time-specific gene targeting in the mouse. Methods. 2001; 24:71–80.
Article6. Ko SO, Chung IH, Xu X, Oka S, Zhao H, Cho ES, et al. Smad4 is required to regulate the fate of cranial neural crest cells. Dev Biol. 2007; 312:435–47.
Article7. Lan Y, Ovitt CE, Cho ES, Maltby KM, Wang Q, Jiang R. Odd-skipped related 2 (Osr2) encodes a key intrinsic regulator of secondary palate growth and morphogenesis. Development. 2004; 131:3207–16.
Article8. Lan Y, Jiang R. Sonic hedgehog signaling regulates reciprocal epithelial-mesenchymal interactions controlling palatal outgrowth. Development. 2009; 136:1387–96.
Article9. Yang X, Li C, Herrera PL, Deng CX. Generation of Smad4/Dpc4 conditional knockout mice. Genesis. 2002; 32:80–1.10. Martin SJ, Newmeyer DD, Mathias S, Farschon DM, Wang HG, Reed JC, et al. Cell-free reconstitution of Fas-, UV radiation- and ceramide-induced apoptosis. EMBO J. 1995; 14:5191–200.
Article11. Ferguson MWJ, Honig LS. Epithelial-mesenchymal interactions during vertebrate palatogenesis. Curr Top Dev Biol. 1984; 19:137–64.12. Dudas M, Kim J, Li WY, Nagy A, Larsson J, Karlsson S, et al. Epithelial and ectomesenchymal role of the type I TGF-βrecep-tor ALK5 during facial morphogenesis and palatal fusion. Dev Biol. 2006; 296:298–314.13. Kaartinen V, Voncken JW, Shuler C, Warburton D, Bu D, Heisterkamp N, et al. Abnormal lung development and cleft palate in mice lacking TGF-β 3 indicates defects of epithelial-mesenchymal interaction. Nat Genet. 1995; 11:415–21.14. Liu W, Sun X, Braut A, Mishina Y, Behringer RR, Mina M, et al. Distinct functions for Bmp signaling in lip and palate fusion in mice. Development. 2005; 132:1453–61.
Article15. Rice R, Spencer-Dene B, Connor EC, Gritli-Linde A, McMahon AP, Dickson C, et al. Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. J Clin Invest. 2004; 113:1692–700.
Article16. Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nature Genet. 1994; 6:348–56.
Article17. Zhao Y, Guo YJ, Tomac AC, Taylor NR, Grinberg A, Lee EJ, et al. Isolated cleft palate in mice with targeted mutation of the LIM homeobox gene Lhx8. Proc Natl Acad Sci U S A. 1999; 96:15002–6.18. Zhang Z, Song Y, Zhao X, Zhang X, Fermin C, Chen Y. Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development. 2002; 129:4135–46.19. Yu L, Gu S, Alappat S, Song Y, Yan M, Zhang X, et al. Shox2-deficient mice exhibit a rare type of incomplete clefting of the secondary palate. Development. 2005; 132:4397–406.
Article20. Ito Y, Yeo JY, Chytil A, Han J, Bringas P Jr, Nakajima A, et al. Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development. 2003; 130:5269–80.21. Xu X, Han J, Ito Y, Bringas P Jr, Urata MM, Chai Y. Cell autonomous requirement for Tgfbr2 in the disappearance of medial edge epithelium during palatal fusion. Dev Biol. 2006; 297:238–48.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Smad4 Mediated TGF-beta/BMP Signaling in Tooth Formation Using Smad4 Conditional Knockout Mouse
- Requirement of Smad4-mediated signaling in odontoblast differentiation and dentin matrix formation
- Altered Expression of Smad Proteins in T or NK-cell Lymphomas
- Clinicopathological Significance of SMAD4 Expression in Breast Cancer
- Smad4 Expression in Gastric Adenocarcinoma