Tuberc Respir Dis.
2006 Feb;60(2):187-193.
Investigation of the Growth Rate Change in Recombinant BCG which was cloned Mycobacterium tuberculosis Adenylate Kinase Mutation Gene or Human Muscle-type Adenylate Kinase Synthetic Gene
- Affiliations
-
- 1Department of Molecular Biology, Korean Institute of Tuberculosis, Seoul, Korea. gbai@hotmail.com
- 2Department of Biochemistry, Hanyang University College of Biotechnology, Ansan, Korea.
Abstract
-
BACKGROUND: Normal cell proliferation and viability is strongly depends on the availability of metabolic energy and the maintenance of the appropriate adenylate-nucleotide pools. Hypothetically, changes in adenylate kinase (AK) expression could therefore be associated with adaptation to altered growth characteristics or inversely altered growth characteristics of proliferating cells could drive the changes in the metabolic profile. This study investigated whether the expression of either AK1 or a Mycobacterium tuberculosis adenylate kinase mutant which has the same catalytic activity of AK1 could affect the growth rate of slow-growing BCG.
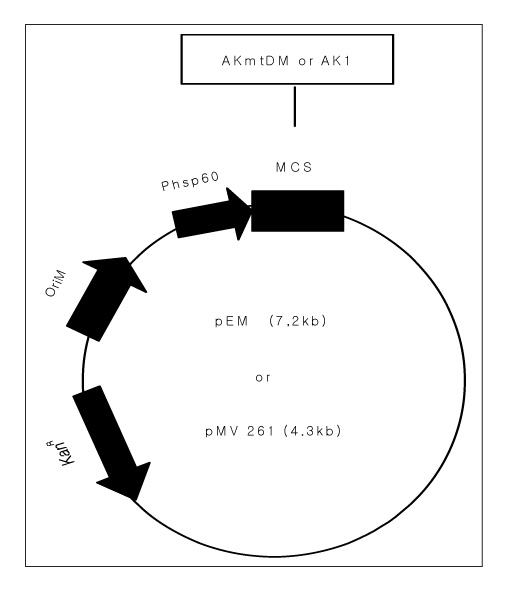
METHOD: Recombinant BCGs, which were cloned the human muscle-type adenylate kinase synthetic gene (AK1) and adenylate kinase mutation gene (AKmtDM) of Mycobacterium tuberculosis into the Mycobacterium/E.coli expression vectors, were constructed. Recombinant BCGs and wild-type BCG were cultured in 7H9 media and the optical density at 600nm was measured at intervals of 2-3 days.
RESULT: There wasn't the growth rate change induced by AK1 or AKmtDM expression in recombinant BCGs.
CONCLUSION
The expression of AK1 or Mycobacterium tuberculosis adenylate kinase mutant in BCG does not affect the growth rate of BCG.
Keyword
MeSH Terms
Figure
Reference
-
1. Dzeja PP, Zeleznikar RJ, Goldberg ND. Adenylate kinase: kinetic behavior in intact cells indicates it is integral to multiple cellular processes. Mol Cell Biochem. 1998. 184:169–182.2. Zeleznikar RJ, Dzeja PP, Goldberg ND. Adenylate kinase-catalyzed phosphoryl transfer couples ATP utilization with its generation by glycolysis in intact muscle. J Biol Chem. 1995. 270:7311–7319.3. van Rompay AR, Johansson M, Karlsson A. Phosphorylation of nucleosides and nucleoside analogues by mammalian nucleoside monophosphate kinases. Pharmacel Ther. 2000. 87:189–198.4. van Rompay AR, Johansson M, Karlsson A. Identification of a novel human adenylate kinase: cDNA cloning, expression analysis, chromosome localization and characterization of the recombinant protein. Eur J Biochem. 1999. 261:509–517.5. Fukami-Kobayashi K, Nosaka M, Nakazawa A, Go M. Ancient divergence of long and short isoforms of adenylate kinase: molecular evolution of the nucleoside monophosphate kinase family. FEBS Lett. 1996. 385:214–220.6. Yoneda T, Sato M, Maeda M, Takagi H. Identification of a novel adenylate kinase system in brain: cloning of the fourth adenylate kinase. Brain Res Mol Brain Res. 1998. 62:187–195.7. Muller CW, Schlauderer GJ, Reinstein J, Schultz GE. Adenylate kinase motions during catalysis: an energetic counterweight balancing substrate binding. Structure. 1996. 4:147–156.8. Muller CW, Schultz GE. Structure of the complex of adenylate kinase from Escherichia coli with the inhibitor P1, P5-di(adenosine-5'-)pentaphosphate. J Mol Biol. 1988. 202:909–912.9. Abele U, Schulz GE. High-resolution structures of adenylate kinase from yeast ligated with inhibitor Ap5A, showing the pathway of phosphoryl transfer. Protein Sci. 1995. 4:1262–1271.10. Wild K, Grafmuller R, Wagner E, Schulz GE. Structure, catalysis and supramolecular assembly of adenylate kinase from maize. Eur J Biochem. 1997. 250:326–331.11. Yan H, Tsai MD. Nucleoside monophosphate kinases: structure, mechanism, and substrate specificity. Adv Enzymol Relat Areas Mol Biol. 1999. 73:103–134.12. Inouye S, Yamada Y, Miura K, Suzuki H, Kawata K, Shinoda K, et al. Distribution and developmental changes of adenylate kinase isozymes in the rat brain: loclization of adenylate kinase 1 in the olfactory bulb. Biochem Biophys Res Commun. 1999. 254:618–622.13. Janssen E, Dzeja PP, Oerlemans F, Simonetti AW, Heerschap A, de Haan A, et al. Adenylate kinase 1 gene deletion disrupts muscle energetic economy despite metabolic rearrangement. EMBO J. 2000. 19:6371–6381.14. Pucar D, Janssen E, Dzeja PP, Juranic N, Macura S, Wieringa B, et al. Compromised energetics in the adenylate kinase AK1 gene knockout heart under metabolic stress. J Biol Chem. 2000. 275:41424–41429.15. Miron S, Munier-Lehmann H, Craescu CT. Structure and dynamic studies on ligand-free adenylate kinase from Mycobacterium tuberculosis revealed a closed conformation that can be related to the reduced catalytic activity. Biochemistry. 2004. 43:67–77.16. Munier-Lehmann H, Burlacu-Miron S, Craescu CT, Mantsch HH, Schultz CP. A new subfamily of short bacterial adenylate kinases with the Mycobacterium tuberculosis enzyme as a model: a predictive and experimental study. Proteins. 1999. 36:238–248.18. de Bruin W, Oerlemans F, Wieringa B. Adenylate kinase I does not affect cellular growth characteristics under normal and metabolic stress conditions. Exp Cell Res. 2004. 297:97–107.19. Markaryan A, Zaborina O, Punj V, Chakrabarty AM. Adenylate kinase as a virulence factor of Pseudomonas aeruginosa. J Bacteriol. 2001. 183:3345–3352.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Protective Efficacy of Recombinant Proteins Adenylate Kinase, Nucleoside Diphosphate Kinase, and Heat-Shock Protein 70 against Mycobacterium tuberculosis Infection in Mice
- Expression of the 38 kDa Protein of Mycobacterium tuberculosis in M . bovis BCG and Use in the Serodiagnosis of Tuberculosis
- Secretion of adenylate kinase 1 is required for extracellular ATP synthesis in C2C12 myotubes
- Serological Kinetics of Adenylate Kinase 3 in Rat Acute Myocardial Infarction
- Stimulation by EGF, bFGF and GnRH of Ovarian Pituitary Adenylate Cyclase-Activating Polypeptide Gene Expression in Cultured Rat Preovulatory Follicles