Immune Netw.
2016 Jun;16(3):176-182. 10.4110/in.2016.16.3.176.
Roles of RUNX1 and PU.1 in CCR3 Transcription
- Affiliations
-
- 1Department of Bionano Technology, Hanyang University, Ansan 15588, Korea. iychu@hanyang.ac.kr
- 2Division of Molecular and Life Sciences, College of Science and Technology, Hanyang University, Ansan 15588, Korea.
- 3Department of Bioscience and Biotechnology and Protein Research Center of GRRC, College of Natural Sciences, Hankuk University of Foreign Studies, Yongin 17035, Korea.
- KMID: 2299791
- DOI: http://doi.org/10.4110/in.2016.16.3.176
Abstract
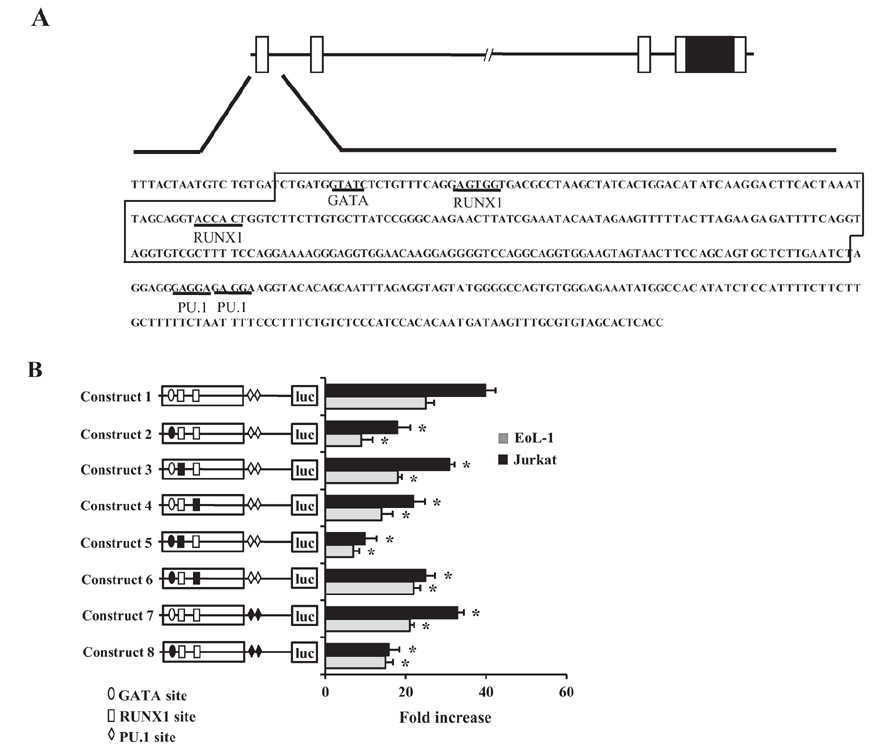
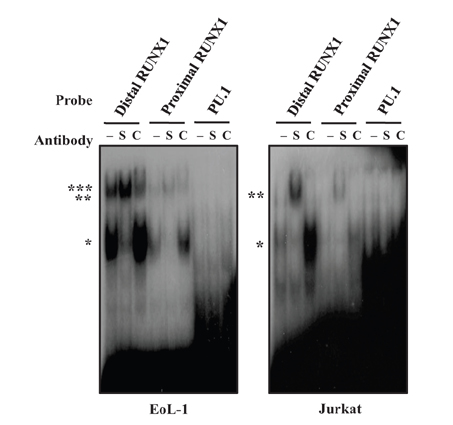
- CCR3 is a chemokine receptor that mediates the accumulation of allergic inflammatory cells, including eosinophils and Th2 cells, at inflamed sites. The regulatory sequence of the CCR3 gene, contains two Runt-related transcription factor (RUNX) 1 sites and two PU.1 sites, in addition to a functional GATA site for transactivation of the CCR3 gene. In the present study, we examined the effects of the cis-acting elements of RUNX1 and PU.1 on transcription of the gene in EoL-1 eosinophilic cells and Jurkat T cells, both of which expressed functional surface CCR3 and these two transcription factors. Introduction of RUNX1 siRNA or PU.1 siRNA resulted in a modest decrease in CCR3 reporter activity in both cell types, compared with transfection of GATA-1 siRNA. Cotransfection of the two siRNAs led to inhibition in an additive manner. EMSA analysis showed that RUNX1, in particular, bound to its binding motifs. Mutagenesis analysis revealed that all point mutants lacking RUNX1- and PU.1-binding sites exhibited reduced reporter activities. These results suggest that RUNX1 and PU.1 participate in transcriptional regulation of the CCR3 gene.
Keyword
MeSH Terms
Figure
Reference
-
1. Pease JE, Williams TJ. Chemokines and their receptors in allergic disease. J Allergy Clin Immunol. 2006; 118:305–318.
Article2. Rothenberg ME, Hogan SP. The eosinophil. Annu Rev Immunol. 2006; 24:147–174.
Article3. Willems LI, Ijzerman AP. Small molecule antagonists for chemokine CCR3 receptors. Med Res Rev. 2010; 30:778–817.
Article4. Mori Y, Iwasaki H, Kohno K, Yoshimoto G, Kikushige Y, Okeda A, Uike N, Niiro H, Takenaka K, Nagafuji K, Miyamoto T, Harada M, Takatsu K, Akashi K. Identification of the human eosinophil lineage-committed progenitor: revision of phenotypic definition of the human common myeloid progenitor. J Exp Med. 2009; 206:183–193.
Article5. Voehringer D, van RN, Locksley RM. Eosinophils develop in distinct stages and are recruited to peripheral sites by alternatively activated macrophages. J Leukoc Biol. 2007; 81:1434–1444.
Article6. Kim BS, Uhm TG, Lee SK, Lee SH, Kang JH, Park CS, Chung IY. The crucial role of GATA-1 in CCR3 gene transcription: modulated balance by multiple GATA elements in the CCR3 regulatory region. J Immunol. 2010; 185:6866–6875.
Article7. Vijh S, Dayhoff DE, Wang CE, Imam Z, Ehrenberg PK, Michael NL. Transcription regulation of human chemokine receptor CCR3: evidence for a rare TATA-less promoter structure conserved between drosophila and humans. Genomics. 2002; 80:86–95.
Article8. Zimmermann N, Daugherty BL, Kavanaugh JL, El-Awar FY, Moulton EA, Rothenberg ME. Analysis of the CC chemokine receptor 3 gene reveals a complex 5' exon organization, a functional role for untranslated exon 1, and a broadly active promoter with eosinophil-selective elements. Blood. 2000; 96:2346–2354.
Article9. Scotet E, Schroeder S, Lanzavecchia A. Molecular regulation of CC-chemokine receptor 3 expression in human T helper 2 cells. Blood. 2001; 98:2568–2570.
Article10. Klemsz MJ, McKercher SR, Celada A, Van BC, Maki RA. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990; 61:113–124.
Article11. Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994; 265:1573–1577.
Article12. McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, Klemsz M, Feeney AJ, Wu GE, Paige CJ, Maki RA. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996; 15:5647–5658.
Article13. DeKoter RP, Walsh JC, Singh H. PU.1 regulates both cytokine-dependent proliferation and differentiation of granulocyte/macrophage progenitors. EMBO J. 1998; 17:4456–4468.
Article14. McNagny KM, Sieweke MH, Doderlein G, Graf T, Nerlov C. Regulation of eosinophil-specific gene expression by a C/EBP-Ets complex and GATA-1. EMBO J. 1998; 17:3669–3680.
Article15. Kodandapani R, Pio F, Ni CZ, Piccialli G, Klemsz M, McKercher S, Maki RA, Ely KR. A new pattern for helix-turn-helix recognition revealed by the PU.1 ETSdomain-DNA complex. Nature. 1996; 380:456–460.
Article16. Growney JD, Shigematsu H, Li Z, Lee BH, Adelsperger J, Rowan R, Curley DP, Kutok JL, Akashi K, Williams IR, Speck NA, Gilliland DG. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005; 106:494–504.
Article17. Collins A, Littman DR, Taniuchi I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat Rev Immunol. 2009; 9:106–115.
Article18. Mukai K, BenBarak MJ, Tachibana M, Nishida K, Karasuyama H, Taniuchi I, Galli SJ. Critical role of P1-Runx1 in mouse basophil development. Blood. 2012; 120:76–85.
Article19. Meyers S, Downing JR, Hiebert SW. Identification of AML-1 and the (8;21) translocation protein (AML-1/ETO) as sequence-specific DNA-binding proteins: the runt homology domain is required for DNA binding and protein-protein interactions. Mol Cell Biol. 1993; 13:6336–6345.
Article20. Kong SK, Kim BS, Uhm TG, Lee W, Lee GR, Park CS, Lee CH, Chung IY. Different GATA factors dictate CCR3 transcription in allergic inflammatory cells in a cell type-specific manner. J Immunol. 2013; 190:5747–5756.
Article21. Elagib KE, Racke FK, Mogass M, Khetawat R, Delehanty LL, Goldfarb AN. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood. 2003; 101:4333–4341.
Article22. Anderson KL, Smith KA, Pio F, Torbett BE, Maki RA. Neutrophils deficient in PU.1 do not terminally differentiate or become functionally competent. Blood. 1998; 92:1576–1585.
Article23. Du J, Stankiewicz MJ, Liu Y, Xi Q, Schmitz JE, Lekstrom-Himes JA, Ackerman SJ. Novel combinatorial interactions of GATA-1, PU.1, and C/EBPε isoforms regulate transcription of the gene encoding eosinophil granule major basic protein. J Biol Chem. 2002; 277:43481–43494.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Eosinophil Development, Regulation of Eosinophil-Specific Genes, and Role of Eosinophils in the Pathogenesis of Asthma
- RUNX1 Germline Mutation in a Patient with Chronic Thrombocytopenia
- CpG methylation at GATA elements in the regulatory region of CCR3 positively correlates with CCR3 transcription
- MicroRNA-181a-5p Curbs Osteogenic Differentiation and Bone Formation Partially Through Impairing Runx1-Dependent Inhibition of AIF-1 Transcription
- The Macrophage-Specific Transcription Factor Can Be Modified Posttranslationally by Ubiquitination in the Lipopolysaccharide-Treated Macrophages