J Breast Cancer.
2012 Jun;15(2):230-238. 10.4048/jbc.2012.15.2.230.
Development of Novel Breast Cancer Recurrence Prediction Model Using Support Vector Machine
- Affiliations
-
- 1Department of Biomedical Informatics, Ajou University School of Medicine, Suwon, Korea. veritas@ajou.ac.kr
- 2Department of Surgery, Ajou University School of Medicine, Suwon, Korea.
- 3Department of Surgery, Samsung Medical Center, Seoul, Korea.
- 4Department of Surgery, Seoul National University College of Medicine, Seoul, Korea.
- KMID: 2242195
- DOI: http://doi.org/10.4048/jbc.2012.15.2.230
Abstract
- PURPOSE
The prediction of breast cancer recurrence is a crucial factor for successful treatment and follow-up planning. The principal objective of this study was to construct a novel prognostic model based on support vector machine (SVM) for the prediction of breast cancer recurrence within 5 years after breast cancer surgery in the Korean population, and to compare the predictive performance of the model with the previously established models.
METHODS
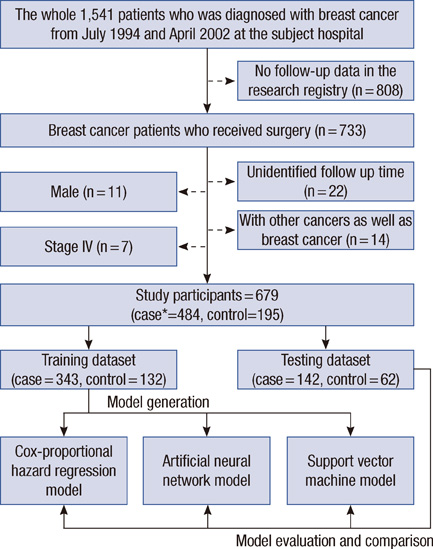
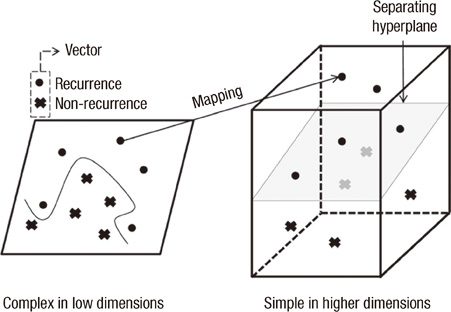
Data on 679 patients, who underwent breast cancer surgery between 1994 and 2002, were collected retrospectively from a Korean tertiary teaching hospital. The following variables were selected as independent variables for the prognostic model, by using the established medical knowledge and univariate analysis: histological grade, tumor size, number of metastatic lymph node, estrogen receptor, lymphovascular invasion, local invasion of tumor, and number of tumors. Three prediction algorithms, with each using SVM, artificial neural network and Cox-proportional hazard regression model, were constructed and compared with one another. The resultant and most effective model based on SVM was compared with previously established prognostic models, which included Adjuvant! Online, Nottingham prognostic index (NPI), and St. Gallen guidelines.
RESULTS
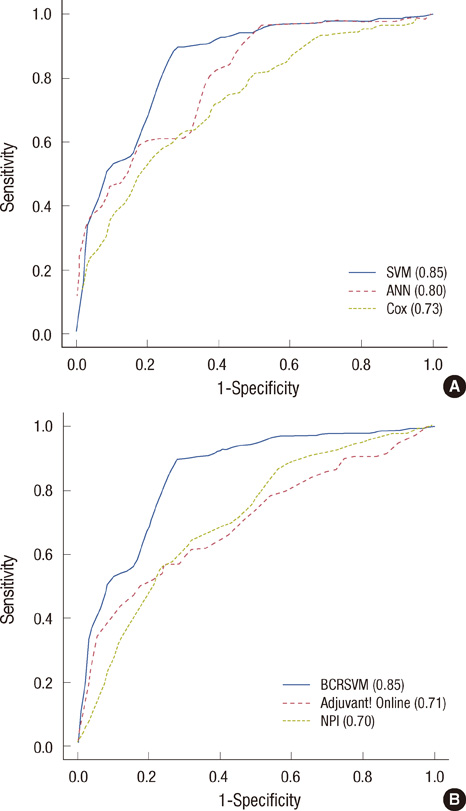
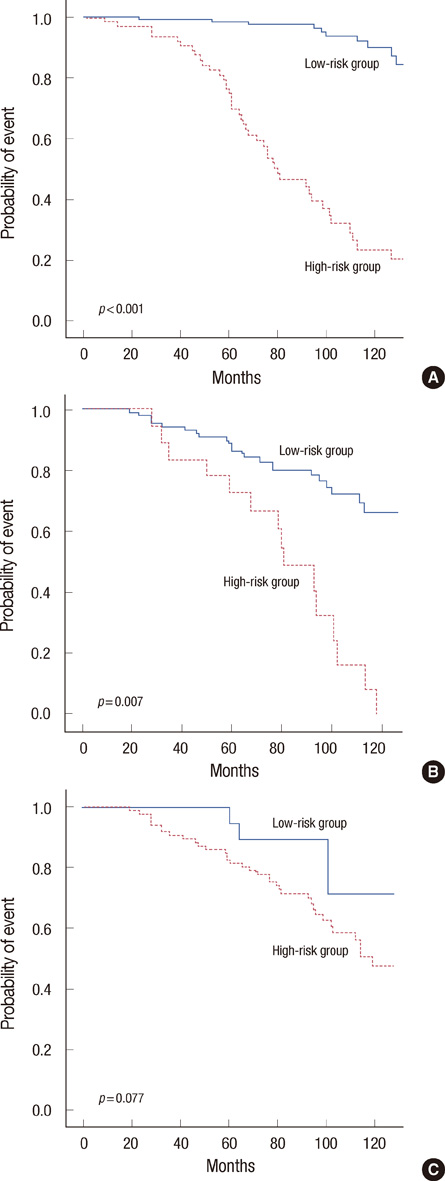
The SVM-based prediction model, named 'breast cancer recurrence prediction based on SVM (BCRSVM),' proposed herein outperformed other prognostic models (area under the curve=0.85, 0.71, 0.70, respectively for the BCRSVM, Adjuvant! Online, and NPI). The BCRSVM evidenced substantially high sensitivity (0.89), specificity (0.73), positive predictive values (0.75), and negative predictive values (0.89).
CONCLUSION
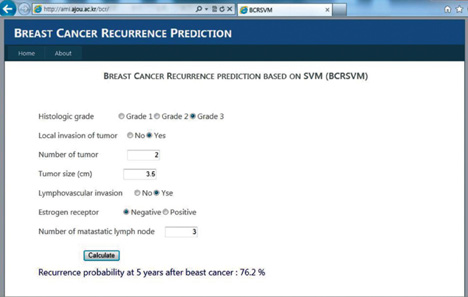
As the selected prognostic factors can be easily obtained in clinical practice, the proposed model might prove useful in the prediction of breast cancer recurrence. The prediction model is freely available in the website (http://ami.ajou.ac.kr/bcr/).
MeSH Terms
Figure
Cited by 1 articles
-
Nomogram of Naive Bayesian Model for Recurrence Prediction of Breast Cancer
Woojae Kim, Ku Sang Kim, Rae Woong Park
Healthc Inform Res. 2016;22(2):89-94. doi: 10.4258/hir.2016.22.2.89.
Reference
-
1. Na KY, Kim KS, Lee JE, Kim HJ, Yang JH, Ahn SH, et al. The 70-gene prognostic signature for Korean breast cancer patients. J Breast Cancer. 2011. 14:33–38.
Article2. Muñoz M, Estévez LG, Alvarez I, Fernández Y, Margelí M, Tusquets I, et al. Evaluation of international treatment guidelines and prognostic tests for the treatment of early breast cancer. Cancer Treat Rev. 2008. 34:701–709.
Article3. Goldhirsch A, Wood WC, Gelber RD, Coates AS, Thürlimann B, Senn HJ, et al. Progress and promise: highlights of the international expert consensus on the primary therapy of early breast cancer 2007. Ann Oncol. 2007. 18:1133–1144.
Article4. Goldhirsch A, Ingle JN, Gelber RD, Coates AS, Thürlimann B, Senn HJ, et al. Thresholds for therapies: highlights of the St Gallen International Expert Consensus on the primary therapy of early breast cancer 2009. Ann Oncol. 2009. 20:1319–1329.
Article5. Iwamoto E, Fukutomi T, Akashi-Tanaka S. Validation and problems of St-Gallen recommendations of adjuvant therapy for node-negative invasive breast cancer in Japanese patients. Jpn J Clin Oncol. 2001. 31:259–262.
Article6. Jung SY, Han W, Lee JW, Ko E, Kim E, Yu JH, et al. Ki-67 expression gives additional prognostic information on St. Gallen 2007 and Adjuvant! Online risk categories in early breast cancer. Ann Surg Oncol. 2009. 16:1112–1121.
Article7. Boyages J, Chua B, Taylor R, Bilous M, Salisbury E, Wilcken N, et al. Use of the St Gallen classification for patients with node-negative breast cancer may lead to overuse of adjuvant chemotherapy. Br J Surg. 2002. 89:789–796.
Article8. Roila F, Ballatori E, Patoia L, Palazzo S, Veronesi A, Frassoldati A, et al. Adjuvant systemic therapies in women with breast cancer: an audit of clinical practice in Italy. Ann Oncol. 2003. 14:843–848.
Article9. Buyse M, Loi S, van't Veer L, Viale G, Delorenzi M, Glas AM, et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006. 98:1183–1192.
Article10. van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002. 347:1999–2009.
Article11. Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009. 360:790–800.
Article12. Galea MH, Blamey RW, Elston CE, Ellis IO. The Nottingham prognostic index in primary breast cancer. Breast Cancer Res Treat. 1992. 22:207–219.
Article13. Ravdin PM, Siminoff LA, Davis GJ, Mercer MB, Hewlett J, Gerson N, et al. Computer program to assist in making decisions about adjuvant therapy for women with early breast cancer. J Clin Oncol. 2001. 19:980–991.
Article14. Jerez JM, Franco L, Alba E, Llombart-Cussac A, Lluch A, Ribelles N, et al. Improvement of breast cancer relapse prediction in high risk intervals using artificial neural networks. Breast Cancer Res Treat. 2005. 94:265–272.
Article15. Jerez-Aragones JM, Gomez-Ruiz JA, Ramos-Jimenez G, Munoz-Perez J, Alba-Conejo E. A combined neural network and decision trees model for prognosis of breast cancer relapse. Artif Intell Med. 2003. 27:45–63.
Article16. Sargent DJ. Comparison of artificial neural networks with other statistical approaches: results from medical data sets. Cancer. 2001. 91:8 Suppl. 1636–1642.
Article17. Lisboa PJ, Wong H, Harris P, Swindell R. A Bayesian neural network approach for modelling censored data with an application to prognosis after surgery for breast cancer. Artif Intell Med. 2003. 28:1–25.
Article18. Olivotto IA, Bajdik CD, Ravdin PM, Speers CH, Coldman AJ, Norris BD, et al. Population-based validation of the prognostic model Adjuvant! for early breast cancer. J Clin Oncol. 2005. 23:2716–2725.
Article19. Aitkin M, Laird N, Francis B. A reanalysis of the Stanford heart transplant data. J Am Stat Assoc. 1983. 78:264–274.
Article20. Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995. 20:273–297.
Article21. Meyer D, Leisch F, Hornik K. The support vector machine under test. Neurocomputing. 2003. 55:169–186.
Article22. Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. Mach Learn. 2002. 46:389–422.23. Furey TS, Cristianini N, Duffy N, Bednarski DW, Schummer M, Haussler D. Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics. 2000. 16:906–914.
Article24. Estévez PA, Tesmer M, Perez CA, Zurada JM. Normalized mutual information feature selection. IEEE Trans Neural Netw. 2009. 20:189–201.
Article25. Moody J, Darken CJ. Fast learning in networks of locally-tuned processing units. Neural Comput. 1989. 1:281–294.
Article26. Kuncheva LI, Hadjitodorov ST. Using diversity in cluster ensembles. 2004. 2:In : 2004 IEEE International Conference on Systems, Man and Cybernetics; 1214–1219.27. Butte AJ, Kohane IS, Kohane IS. Mutual information relevance networks: functional genomic clustering using pairwise entropy measurements. Pac Symp Biocomput. 2000. 5:415–426.
Article28. Ishitobi M, Goranova TE, Komoike Y, Motomura K, Koyama H, Glas AM, et al. Clinical utility of the 70-gene MammaPrint profile in a Japanese population. Jpn J Clin Oncol. 2010. 40:508–512.
Article29. Sun JM, Han W, Im SA, Kim TY, Park IA, Noh DY, et al. A combination of HER-2 status and the St. Gallen classification provides useful information on prognosis in lymph node-negative breast carcinoma. Cancer. 2004. 101:2516–2522.
Article30. Tutt A, Wang A, Rowland C, Gillett C, Lau K, Chew K, et al. Risk estimation of distant metastasis in node-negative, estrogen receptor-positive breast cancer patients using an RT-PCR based prognostic expression signature. BMC Cancer. 2008. 8:339.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Nomogram of Naive Bayesian Model for Recurrence Prediction of Breast Cancer
- Comparison of survival prediction models for pancreatic cancer: Cox model versus machine learning models
- An improved machine learning model for calculation of intraocular lens power during cataract surgery in Republic of Korea: development
- Development and External Validation of a Machine Learning Model to Predict Pathological Complete Response After Neoadjuvant Chemotherapy in Breast Cancer
- A Comparison of Intensive Care Unit Mortality Prediction Models through the Use of Data Mining Techniques