J Korean Neurosurg Soc.
2016 Mar;59(2):83-90. 10.3340/jkns.2016.59.2.83.
The Similarities and Differences between Intracranial and Spinal Ependymomas : A Review from a Genetic Research Perspective
- Affiliations
-
- 1Department of Neurosurgery, Ilsan Paik Hospital, Inje University College of Medicine, Goyang, Korea.
- 2Department of Neurosurgery, Seoul National University Hospital, Seoul, Korea. chungc@snu.ac.kr
- 3Department of Neurosurgery, Seoul National University College of Medicine, Seoul, Korea.
- 4Neuroscience Research Institute, Seoul National University Medical Research Center, Seoul, Korea.
- 5Clinical Research Institute, Seoul National University Hospital, Seoul, Korea.
- 6Department of Brain and Cognitive Sciences, Seoul National University College of Natural Sciences, Seoul, Korea.
- 7Bioinformatics, Samsung Gene Institute, Samsung Medical Center, Seoul, Korea.
- KMID: 2192036
- DOI: http://doi.org/10.3340/jkns.2016.59.2.83
Abstract
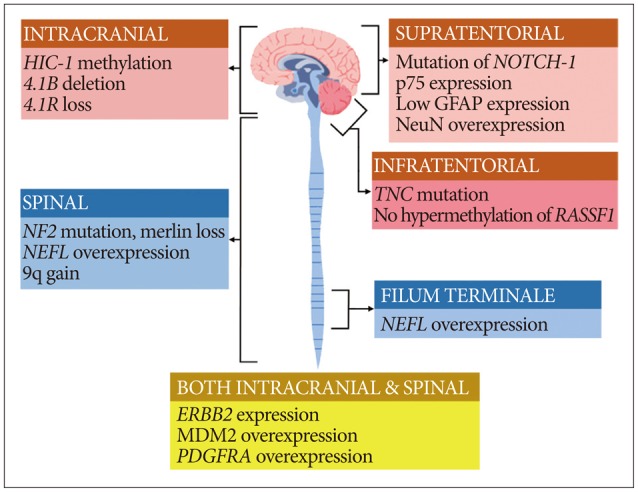
- Ependymomas occur in both the brain and spine. The prognosis of these tumors sometimes differs for different locations. The genetic landscape of ependymoma is very heterogeneous despite the similarity of histopathologic findings. In this review, we describe the genetic differences between spinal ependymomas and their intracranial counterparts to better understand their prognosis. From the literature review, many studies have reported that spinal cord ependymoma might be associated with NF2 mutation, NEFL overexpression, Merlin loss, and 9q gain. In myxopapillary ependymoma, NEFL and HOXB13 overexpression were reported to be associated. Prior studies have identified HIC-1 methylation, 4.1B deletion, and 4.1R loss as common features in intracranial ependymoma. Supratentorial ependymoma is usually characterized by NOTCH-1 mutation and p75 expression. TNC mutation, no hypermethylation of RASSF1A, and GFAP/NeuN expression may be diagnostic clues of posterior fossa ependymoma. Although MEN1, TP53, and PTEN mutations are rarely reported in ependymoma, they may be related to a poor prognosis, such as recurrence or metastasis. Spinal ependymoma has been found to be quite different from intracranial ependymoma in genetic studies, and the favorable prognosis in spinal ependymoma may be the result of the genetic differences. A more detailed understanding of these various genetic aberrations may enable the identification of more specific prognostic markers as well as the development of customized targeted therapies.
Keyword
MeSH Terms
Figure
Reference
-
1. Ahmad ZK, Brown CM, Cueva RA, Ryan AF, Doherty JK. ErbB expression, activation, and inhibition with lapatinib and tyrphostin (AG825) in human vestibular schwannomas. Otol Neurotol. 2011; 32:841–847. PMID: 21659924.
Article2. Andreiuolo F, Puget S, Peyre M, Dantas-Barbosa C, Boddaert N, Philippe C, et al. Neuronal differentiation distinguishes supratentorial and infratentorial childhood ependymomas. Neuro Oncol. 2010; 12:1126–1134. PMID: 20615923.
Article3. Athanasiou A, Perunovic B, Quilty RD, Gorgoulis VG, Kittas C, Love S. Expression of mos in ependymal gliomas. Am J Clin Pathol. 2003; 120:699–705. PMID: 14608895.
Article4. Barton VN, Donson AM, Kleinschmidt-DeMasters BK, Birks DK, Handler MH, Foreman NK. Unique molecular characteristics of pediatric myxopapillary ependymoma. Brain Pathol. 2010; 20:560–570. PMID: 19793339.
Article5. Bettegowda C, Agrawal N, Jiao Y, Wang Y, Wood LD, Rodriguez FJ, et al. Exomic sequencing of four rare central nervous system tumor types. Oncotarget. 2013; 4:572–583. PMID: 23592488.
Article6. de Bont JM, Packer RJ, Michiels EM, den Boer ML, Pieters R. Biological background of pediatric medulloblastoma and ependymoma : a review from a translational research perspective. Neuro Oncol. 2008; 10:1040–1060. PMID: 18676356.
Article7. Ebert C, von Haken M, Meyer-Puttlitz B, Wiestler OD, Reifenberger G, Pietsch T, et al. Molecular genetic analysis of ependymal tumors. NF2 mutations and chromosome 22q loss occur preferentially in intramedullary spinal ependymomas. Am J Pathol. 1999; 155:627–632. PMID: 10433955.8. Fink KL, Rushing EJ, Schold SC Jr, Nisen PD. Infrequency of p53 gene mutations in ependymomas. J Neurooncol. 1996; 27:111–115. PMID: 8699232.
Article9. Garcia C, Gutmann DH. Nf2/Merlin controls spinal cord neural progenitor function in a Rac1/ErbB2-dependent manner. PLoS One. 2014; 9:e97320. PMID: 24817309.
Article10. Gilbertson RJ, Bentley L, Hernan R, Junttila TT, Frank AJ, Haapasalo H, et al. ERBB receptor signaling promotes ependymoma cell proliferation and represents a potential novel therapeutic target for this disease. Clin Cancer Res. 2002; 8:3054–3064. PMID: 12374672.11. Gonzalez-Gomez P, Bello MJ, Alonso ME, Arjona D, Lomas J, de Campos JM, et al. CpG island methylation status and mutation analysis of the RB1 gene essential promoter region and protein-binding pocket domain in nervous system tumours. Br J Cancer. 2003; 88:109–114. PMID: 12556968.
Article12. Gupta RK, Sharma MC, Suri V, Kakkar A, Singh M, Sarkar C. Study of chromosome 9q gain, Notch pathway regulators and Tenascin-C in ependymomas. J Neurooncol. 2014; 116:267–274. PMID: 24178439.
Article13. Hagel C, Treszl A, Fehlert J, Harder J, von Haxthausen F, Kern M, et al. Supra- and infratentorial pediatric ependymomas differ significantly in NeuN, p75 and GFAP expression. J Neurooncol. 2013; 112:191–197. PMID: 23371454.
Article14. Hamilton DW, Lusher ME, Lindsey JC, Ellison DW, Clifford SC. Epigenetic inactivation of the RASSF1A tumour suppressor gene in ependymoma. Cancer Lett. 2005; 227:75–81. PMID: 16051033.
Article15. Huang B, Starostik P, Kühl J, Tonn JC, Roggendorf W. Loss of heterozygosity on chromosome 22 in human ependymomas. Acta Neuropathol. 2002; 103:415–420. PMID: 11904762.
Article16. Huang B, Starostik P, Schraut H, Krauss J, Sörensen N, Roggendorf W. Human ependymomas reveal frequent deletions on chromosomes 6 and 9. Acta Neuropathol. 2003; 106:357–362. PMID: 12898154.
Article17. Johnson RA, Wright KD, Poppleton H, Mohankumar KM, Finkelstein D, Pounds SB, et al. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature. 2010; 466:632–636. PMID: 20639864.
Article18. Kilday JP, Rahman R, Dyer S, Ridley L, Lowe J, Coyle B, et al. Pediatric ependymoma : biological perspectives. Mol Cancer Res. 2009; 7:765–786. PMID: 19531565.19. Korshunov A, Witt H, Hielscher T, Benner A, Remke M, Ryzhova M, et al. Molecular staging of intracranial ependymoma in children and adults. J Clin Oncol. 2010; 28:3182–3190. PMID: 20516456.
Article20. Kraus JA, de Millas W, Sörensen N, Herbold C, Schichor C, Tonn JC, et al. Indications for a tumor suppressor gene at 22q11 involved in the pathogenesis of ependymal tumors and distinct from hSNF5/INI1. Acta Neuropathol. 2001; 102:69–74. PMID: 11547953.
Article21. Lamszus K, Lachenmayer L, Heinemann U, Kluwe L, Finckh U, Höppner W, et al. Molecular genetic alterations on chromosomes 11 and 22 in ependymomas. Int J Cancer. 2001; 91:803–808. PMID: 11275983.
Article22. Magrassi L, Conti L, Lanterna A, Zuccato C, Marchionni M, Cassini P, et al. Shc3 affects human high-grade astrocytomas survival. Oncogene. 2005; 24:5198–5206. PMID: 15870690.
Article23. Magrassi L, Marziliano N, Inzani F, Cassini P, Chiaranda I, Skrap M, et al. EDG3 and SHC3 on chromosome 9q22 are co-amplified in human ependymomas. Cancer Lett. 2010; 290:36–42. PMID: 19748727.
Article24. Modena P, Lualdi E, Facchinetti F, Veltman J, Reid JF, Minardi S, et al. Identification of tumor-specific molecular signatures in intracranial ependymoma and association with clinical characteristics. J Clin Oncol. 2006; 24:5223–5233. PMID: 17114655.
Article25. Monoranu CM, Huang B, Zangen IL, Rutkowski S, Vince GH, Gerber NU, et al. Correlation between 6q25.3 deletion status and survival in pediatric intracranial ependymomas. Cancer Genet Cytogenet. 2008; 182:18–26. PMID: 18328946.
Article26. Olsen TK, Gorunova L, Meling TR, Micci F, Scheie D, Due-Tønnessen B, et al. Genomic characterization of ependymomas reveals 6q loss as the most common aberration. Oncol Rep. 2014; 32:483–490. PMID: 24939246.
Article27. Pajtler KW, Witt H, Sill M, Jones DT, Hovestadt V, Kratochwil F, et al. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell. 2015; 27:728–743. PMID: 25965575.
Article28. Parker M, Mohankumar KM, Punchihewa C, Weinlich R, Dalton JD, Li Y, et al. C11orf95-RELA fusions drive oncogenic NF-κB signalling in ependymoma. Nature. 2014; 506:451–455. PMID: 24553141.
Article29. Puget S, Grill J, Valent A, Bieche I, Dantas-Barbosa C, Kauffmann A, et al. Candidate genes on chromosome 9q33-34 involved in the progression of childhood ependymomas. J Clin Oncol. 2009; 27:1884–1892. PMID: 19289631.
Article30. Rajaram V, Gutmann DH, Prasad SK, Mansur DB, Perry A. Alterations of protein 4.1 family members in ependymomas : a study of 84 cases. Mod Pathol. 2005; 18:991–997. PMID: 15731777.
Article31. Rajaram V, Leuthardt EC, Singh PK, Ojemann JG, Brat DJ, Prayson RA, et al. 9p21 and 13q14 dosages in ependymomas. A clinicopathologic study of 101 cases. Mod Pathol. 2004; 17:9–14. PMID: 14631364.
Article32. Reardon DA, Akabani G, Coleman RE, Friedman AH, Friedman HS, Herndon JE 2nd, et al. Salvage radioimmunotherapy with murine iodine-131-labeled antitenascin monoclonal antibody 81C6 for patients with recurrent primary and metastatic malignant brain tumors : phase II study results. J Clin Oncol. 2006; 24:115–122. PMID: 16382120.
Article33. Rogers HA, Kilday JP, Mayne C, Ward J, Adamowicz-Brice M, Schwalbe EC, et al. Supratentorial and spinal pediatric ependymomas display a hypermethylated phenotype which includes the loss of tumor suppressor genes involved in the control of cell growth and death. Acta Neuropathol. 2012; 123:711–725. PMID: 22109108.
Article34. Rubio MP, Correa KM, Ramesh V, MacCollin MM, Jacoby LB, von Deimling A, et al. Analysis of the neurofibromatosis 2 gene in human ependymomas and astrocytomas. Cancer Res. 1994; 54:45–47. PMID: 8261460.35. Scheil S, Brüderlein S, Eicker M, Herms J, Herold-Mende C, Steiner HH, et al. Low frequency of chromosomal imbalances in anaplastic ependymomas as detected by comparative genomic hybridization. Brain Pathol. 2001; 11:133–143. PMID: 11303789.
Article36. Schneider D, Monoranu CM, Huang B, Rutkowski S, Gerber NU, Krauss J, et al. Pediatric supratentorial ependymomas show more frequent deletions on chromosome 9 than infratentorial ependymomas : a microsatellite analysis. Cancer Genet Cytogenet. 2009; 191:90–96. PMID: 19446744.
Article37. Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001; 2:731–737. PMID: 11584300.
Article38. Singh PK, Gutmann DH, Fuller CE, Newsham IF, Perry A. Differential involvement of protein 4.1 family members DAL-1 and NF2 in intracranial and intraspinal ependymomas. Mod Pathol. 2002; 15:526–531. PMID: 12011257.
Article39. Suzuki SO, Iwaki T. Amplification and overexpression of mdm2 gene in ependymomas. Mod Pathol. 2000; 13:548–553. PMID: 10824927.
Article40. Taylor MD, Poppleton H, Fuller C, Su X, Liu Y, Jensen P, et al. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell. 2005; 8:323–335. PMID: 16226707.
Article41. Teo C, Nakaji P, Symons P, Tobias V, Cohn R, Smee R. Ependymoma. Childs Nerv Syst. 2003; 19:270–285. PMID: 12761644.
Article42. Vera-Bolanos E, Aldape K, Yuan Y, Wu J, Wani K, Necesito-Reyes MJ, et al. Clinical course and progression-free survival of adult intracranial and spinal ependymoma patients. Neuro Oncol. 2015; 17:440–447. PMID: 25121770.
Article43. von Haken MS, White EC, Daneshvar-Shyesther L, Sih S, Choi E, Kalra R, et al. Molecular genetic analysis of chromosome arm 17p and chromosome arm 22q DNA sequences in sporadic pediatric ependymomas. Genes Chromosomes Cancer. 1996; 17:37–44. PMID: 8889505.
Article44. Waha A, Koch A, Hartmann W, Mack H, Schramm J, Sörensen N, et al. Analysis of HIC-1 methylation and transcription in human ependymomas. Int J Cancer. 2004; 110:542–549. PMID: 15122586.45. Wang Z, Zhang J, Ye M, Zhu M, Zhang B, Roy M, et al. Tumor suppressor role of protein 4.1B/DAL-1. Cell Mol Life Sci. 2014; 71:4815–4830. PMID: 25183197.
Article46. Wani K, Armstrong TS, Vera-Bolanos E, Raghunathan A, Ellison D, Gilbertson R, et al. A prognostic gene expression signature in infratentorial ependymoma. Acta Neuropathol. 2012; 123:727–738. PMID: 22322993.
Article47. Ward S, Harding B, Wilkins P, Harkness W, Hayward R, Darling JL, et al. Gain of 1q and loss of 22 are the most common changes detected by comparative genomic hybridisation in paediatric ependymoma. Genes Chromosomes Cancer. 2001; 32:59–66. PMID: 11477662.
Article48. Witt H, Mack SC, Ryzhova M, Bender S, Sill M, Isserlin R, et al. Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell. 2011; 20:143–157. PMID: 21840481.
Article49. Yang I, Nagasawa DT, Kim W, Spasic M, Trang A, Lu DC, et al. Chromosomal anomalies and prognostic markers for intracranial and spinal ependymomas. J Clin Neurosci. 2012; 19:779–785. PMID: 22516549.
Article50. Zadnik PL, Gokaslan ZL, Burger PC, Bettegowda C. Spinal cord tumours : advances in genetics and their implications for treatment. Nat Rev Neurol. 2013; 9:257–266. PMID: 23528542.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Case of Giant Ependymomas of the Cauda Equina associated with Thoracic Intradural Spinal A-V Malformations
- Clinicoradiologic Characteristics of Intradural Extramedullary Conventional Spinal Ependymoma
- Acute Paraplegia as a Result of Hemorrhagic Spinal Ependymoma Masked by Spinal Anesthesia: Case Report and Review of Literature
- Non-Enhancing Intradural Extramedullary Ependymoma: A Case Report
- Intramedullary Ependymoma in the Spinal Cord: A Report of Two Cases