J Clin Neurol.
2014 Jul;10(3):257-261. 10.3988/jcn.2014.10.3.257.
Mutation analysis of SPAST, ATL1, and REEP1 in Korean Patients with Hereditary Spastic Paraplegia
- Affiliations
-
- 1Department of Neurology, Pusan National University School of Medicine, Yangsan, Korea. jhlee.neuro@pusan.ac.kr, dskim@pusan.ac.kr
- 2Research Institute for Convergence of Biomedical Science and Technology, Pusan National University Yangsan Hospital, Yangsan, Korea.
- 3Department of Neurology, Chonnam National University Hospital, Gwangju, Korea.
- 4Department of Neurology, Busan Paik Hospital, Inje University College of Medicine, Busan, Korea.
- KMID: 2179454
- DOI: http://doi.org/10.3988/jcn.2014.10.3.257
Abstract
- BACKGROUND AND PURPOSE
Hereditary spastic paraplegia (HSP) is a genetically heterogeneous group of neurodegenerative disorders that are characterized by progressive spasticity and weakness of the lower limbs. Mutations in the spastin gene (SPAST) are the most common causes of HSP, accounting for 40-67% of autosomal dominant HSP (AD-HSP) and 12-18% of sporadic cases. Mutations in the atlastin-1 gene (ATL1) and receptor expression-enhancing protein 1 gene (REEP1) are the second and third most common causes of AD-HSP, respectively.
METHODS
Direct sequence analysis was used to screen mutations in SPAST, ATL1, and REEP1 in 27 unrelated Korean patients with pure and complicated HSP. Multiplex ligation-dependent probe amplification was also performed to detect copy-number variations of the three genes.
RESULTS
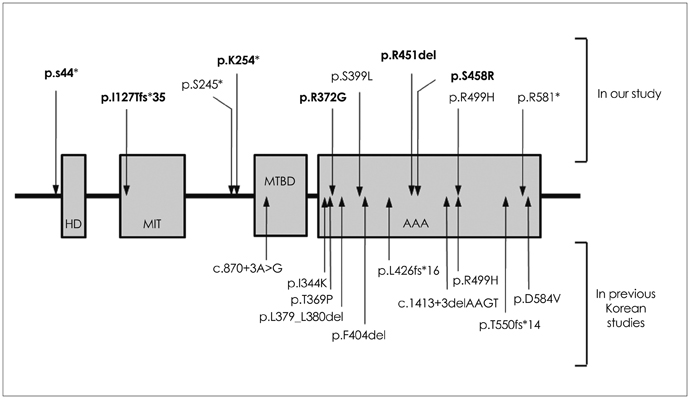
Ten different SPAST mutations were identified in 11 probands, of which the following 6 were novel: c.760A>T, c.131C>A, c.1351_1353delAGA, c.376_377dupTA, c.1114A>G, and c.1372A>C. Most patients with SPAST mutations had AD-HSP (10/11, 91%), and the frequency of SPAST mutations accounted for 66.7% (10/15) of the AD-HSP patients. No significant correlation was found between the presence of the SPAST mutation and any of the various clinical parameters of pure HSP. No ATL1 and REEP1 mutations were detected.
CONCLUSIONS
We conclude that SPAST mutations are responsible for most Korean cases of genetically confirmed AD-HSP. Our observation of the absence of ATL1 and REEP1 mutations needs to be confirmed in larger series.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Pathogenic Variant of REEP1 in a Korean Family with Autosomal-Dominant Hereditary Spastic Paraplegia
Hyung Jun Park, Myung Jun Lee, Jee Eun Lee, Kee Duk Park, Young-Chul Choi
J Clin Neurol. 2018;14(2):248-250. doi: 10.3988/jcn.2018.14.2.248.
Reference
-
1. Salinas S, Proukakis C, Crosby A, Warner TT. Hereditary spastic paraplegia: clinical features and pathogenetic mechanisms. Lancet Neurol. 2008; 7:1127–1138.
Article2. Depienne C, Stevanin G, Brice A, Durr A. Hereditary spastic paraplegias: an update. Curr Opin Neurol. 2007; 20:674–680.
Article3. Finsterer J, Löscher W, Quasthoff S, Wanschitz J, Auer-Grumbach M, Stevanin G. Hereditary spastic paraplegias with autosomal dominant, recessive, X-linked, or maternal trait of inheritance. J Neurol Sci. 2012; 318:1–18.
Article4. Fink JK. Hereditary spastic paraplegia. Curr Neurol Neurosci Rep. 2006; 6:65–76.
Article5. Depienne C, Tallaksen C, Lephay JY, Bricka B, Poea-Guyon S, Fontaine B, et al. Spastin mutations are frequent in sporadic spastic paraparesis and their spectrum is different from that observed in familial cases. J Med Genet. 2006; 43:259–265.
Article6. Crippa F, Panzeri C, Martinuzzi A, Arnoldi A, Redaelli F, Tonelli A, et al. Eight novel mutations is SPG4 in a large sample of patients with hereditary spastic paraplegia. Arch Neurol. 2006; 63:750–755.
Article7. Park SY, Ki CS, Kim HJ, Kim JW, Sung DH, Kim BJ, et al. Mutation analysis of SPG4 and SPG3A genes and its implication in molecular diagnosis of Korean patients with hereditary spastic paraplegia. Arch Neurol. 2005; 62:1118–1121.
Article8. McCorquodale DS 3rd, Ozomaro U, Huang J, Montenegro G, Kushman A, Citrigno L, et al. Mutation screening of spastin, atlastin, and REEP1 in hereditary spastic paraplegia. Clin Genet. 2011; 79:523–530.
Article9. Lim JS, Sung JJ, Hong YH, Park SS, Park KS, Cha JI, et al. A novel splicing mutation (c.870+3A>G) in SPG4 in a Korean family with hereditary spastic paraplegia. J Neurol Sci. 2010; 290:186–189.
Article10. Yi SE, Hong YH, Kim DH, Lee JS, Kim GH, Yoo HW, et al. Autosomal dominant hereditary spastic paraplegia relavant with a novel Thr369Pro mutation in SPAST gene. J Korean Neurol Assoc. 2011; 29:365–367.11. Kwon MJ, Lee ST, Kim JW, Sung DH, Ki CS. Clinical and genetic analysis of a Korean family with hereditary spastic paraplegia type 3. Ann Clin Lab Sci. 2010; 40:375–379.12. Hazan J, Fonknechten N, Mavel D, Paternotte C, Samson D, Artiguenave F, et al. Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat Genet. 1999; 23:296–303.
Article13. Zhao X, Alvarado D, Rainier S, Lemons R, Hedera P, Weber CH, et al. Mutations in a newly identified GTPase gene cause autosomal dominant hereditary spastic paraplegia. Nat Genet. 2001; 29:326–331.
Article14. Harding AE. Classification of the hereditary ataxias and paraplegias. Lancet. 1983; 1:1151–1155.
Article15. Fonknechten N, Mavel D, Byrne P, Davoine CS, Cruaud C, Bönsch D, et al. Spectrum of SPG4 mutations in autosomal dominant spastic paraplegia. Hum Mol Genet. 2000; 9:637–644.
Article16. Lindsey JC, Lusher ME, McDermott CJ, White KD, Reid E, Rubinsztein DC. Mutation analysis of the spastin gene (SPG4) in patients with hereditary spastic paraparesis. J Med Genet. 2000; 37:759–765.
Article17. Guelly C, Zhu PP, Leonardis L, Papić L, Zidar J, Schabhüttl M, et al. Targeted high-throughput sequencing identifies mutations in atlastin-1 as a cause of hereditary sensory neuropathy type 1. Am J Hum Genet. 2011; 88:99–105.
Article18. Du J, Shen L, Zhao GH, Wang YG, Liao SS, Chen C, et al. Receptor expression-enhancing protein 1 gene (SPG31) mutations are rare in Chinese Han patients with hereditary spastic paraplegia. Chin Med J (Engl). 2009; 122:2064–2066.19. Patrono C, Scarano V, Cricchi F, Melone MA, Chiriaco M, Napolitano A, et al. Autosomal dominant hereditary spastic paraplegia: DHPLC-based mutation analysis of SPG4 reveals eleven novel mutations. Hum Mutat. 2005; 25:506.
Article20. Meijer IA, Hand CK, Cossette P, Figlewicz DA, Rouleau GA. Spectrum of SPG4 mutations in a large collection of North American families with hereditary spastic paraplegia. Arch Neurol. 2002; 59:281–286.
Article21. Sauter S, Miterski B, Klimpe S, Bönsch D, Schöls L, Visbeck A, et al. Mutation analysis of the spastin gene (SPG4) in patients in Germany with autosomal dominant hereditary spastic paraplegia. Hum Mutat. 2002; 20:127–132.
Article22. Yabe I, Sasaki H, Tashiro K, Matsuura T, Takegami T, Satoh T. Spastin gene mutation in Japanese with hereditary spastic paraplegia. J Med Genet. 2002; 39:e46.
Article23. Tang B, Zhao G, Xia K, Pan Q, Luo W, Shen L, et al. Three novel mutations of the spastin gene in Chinese patients with hereditary spastic paraplegia. Arch Neurol. 2004; 61:49–55.
Article24. Beetz C, Nygren AO, Schickel J, Auer-Grumbach M, Bürk K, Heide G, et al. High frequency of partial SPAST deletions in autosomal dominant hereditary spastic paraplegia. Neurology. 2006; 67:1926–1930.
Article25. Depienne C, Fedirko E, Forlani S, Cazeneuve C, Ribaï P, Feki I, et al. Exon deletions of SPG4 are a frequenct cause of hereditary spastic paraplegia. J Med Genet. 2007; 44:281–284.
Article26. Züchner S, Wang G, Tran-Viet KN, Nance MA, Gaskell PC, Vance JM, et al. Mutations in the novel mitochondrial protein REEP1 cause hereditary spastic paraplegia type 31. Am J Hum Genet. 2006; 79:365–369.
Article27. Beetz C, Schüle R, Deconinck T, Tran-Viet KN, Zhu H, Kremer BP, et al. REEP1 mutation spectrum and genotype/phenotype correlation in hereditary spastic paraplegia type 31. Brain. 2008; 131(Pt 4):1078–1086.
Article28. Battini R, Fogli A, Borghetti D, Michelucci A, Perazza S, Baldinotti F, et al. Clinical and genetic findings in a series of Italian children with pure hereditary spastic paraplegia. Eur J Neurol. 2011; 18:150–157.
Article29. Goizet C, Depienne C, Benard G, Boukhris A, Mundwiller E, Solé G, et al. REEP1 mutations in SPG31: frequency, mutational spectrum, and potential association with mitochondrial morpho-functional dysfunction. Hum Mutat. 2011; 32:1118–1127.
Article30. Sulek A, Elert E, Rajkiewicz M, Zdzienicka E, Stepniak I, Krysa W, et al. Screening for the hereditary spastic paraplegias SPG4 and SPG3A with the multiplex ligation-dependent probe amplification technique in a large population of affected individuals. Neurol Sci. 2013; 34:239–242.
Article31. de Bot ST, van den Elzen RT, Mensenkamp AR, Schelhaas HJ, Willemsen MA, Knoers NV, et al. Hereditary spastic paraplegia due to SPAST mutations in 151 Dutch patients: new clinical aspects and 27 novel mutations. J Neurol Neurosurg Psychiatry. 2010; 81:1073–1078.
Article32. Erichsen AK, Inderhaug E, Mattingsdal M, Eiklid K, Tallaksen CM. Seven novel mutations and four exon deletions in a collection of Norwegian patients with SPG4 hereditary spastic paraplegia. Eur J Neurol. 2007; 14:809–814.
Article33. Magariello A, Muglia M, Patitucci A, Ungaro C, Mazzei R, Gabriele AL, et al. Mutation analysis of the SPG4 gene in Italian patients with pure and complicated forms of spastic paraplegia. J Neurol Sci. 2010; 288:96–100.
Article34. McMonagle P, Byrne PC, Fitzgerald B, Webb S, Parfrey NA, Hutchinson M. Phenotype of AD-HSP due to mutations in the SPAST gene: comparison with AD-HSP without mutations. Neurology. 2000; 55:1794–1800.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Arg460Cys Mutation in SPAST Gene in Patients with Hereditary Spastic Paraplegia
- Autosomal Dominant Hereditary Spastic Paraplegia Relavant with a Novel Thr369Pro Mutation in SPAST Gene
- Erratum: Mutation Analysis of SPAST, ATL1, and REEP1 in Korean Patients with Hereditary Spastic Paraplegia
- Hereditary Spastic Paraplegia with a Novel SPAST Mutation Misdiagnosed with Subacute Combined Degeneration
- Novel Pathogenic Variant of SPAST (c.1413+4A>G) in a Patient with Hereditary Spastic Paraplegia