J Bacteriol Virol.
2012 Dec;42(4):368-371. 10.4167/jbv.2012.42.4.368.
A Novel PA-X Protein Translated from Influenza A Virus Segment 3
- Affiliations
-
- 1Department of Microbiology, College of Medicine, Hallym University, Chuncheon, Gangwon-do, Korea. manseong.park@gmail.com
- 2Center for Medical Science Research, College of Medicine, Hallym University, Chuncheon, Gangwon-do, Korea.
- KMID: 2168675
- DOI: http://doi.org/10.4167/jbv.2012.42.4.368
Abstract
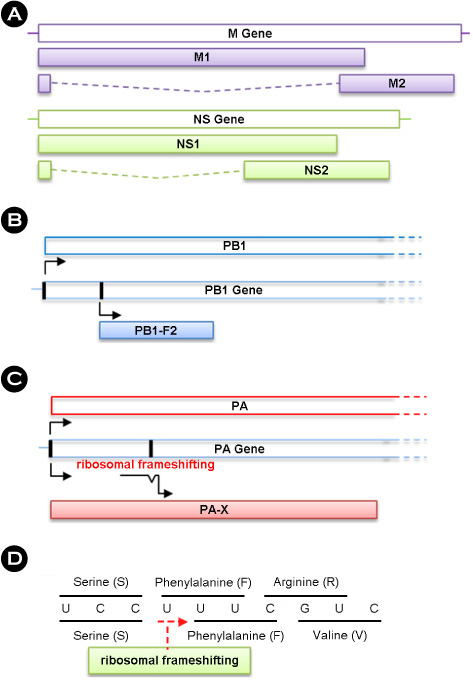
- The pathogenicity of influenza A viruses is a multigenic trait, which is orchestrated by the global networks between eight viral genomic constituents and their cellular interacting partners. A recent report provided information on the finding of a new PA ribosomal frameshifting product, the PA-X protein, in the influenza A virus segment 3, and an endonuclease property was suggested for a possible role of the PA-X protein. In cultured cells, viral growth was not affected by the PA-X protein expression. However, the reduced pathogenicity of mice appeared to be closely associated with the PA-X protein expression. It was also revealed that the PA-X protein was able to modulate host gene expression. Considered together, the PA-X protein can be a cellular signaling modulator and subsequently control viral pathogenicity. By reviewing recent publications, we present new insights in the contribution of the PA-X protein to the cellular signaling network and the resultant viral pathogenicity.
MeSH Terms
Figure
Cited by 1 articles
-
Cell Culture-based Influenza Vaccines as Alternatives to Egg-based Vaccines
Ilseob Lee, Jin Il Kim, Man-Seong Park
J Bacteriol Virol. 2013;43(1):9-17. doi: 10.4167/jbv.2013.43.1.9.
Reference
-
1. Landolt GA, Olsen CW. Up to new tricks - a review of cross-species transmission of influenza A viruses. Anim Health Res Rev. 2007. 8:1–21.
Article2. WHO. Fact sheet on influenza. 2009. Available from: http://www.who.int/mediacentre/factsheets/fs211/en/index.html.3. Klenk HD, Garten W, Matrosovich M. Molecular mechanisms of interspecies transmission and pathogenicity of influenza viruses: Lessons from the 2009 pandemic. Bioessays. 2011. 33:180–188.
Article4. WHO. Cumulative number of confirmed human cases of avian influenza A(H5N1) reported to WHO. 2012. Available from: http://www.who.int/influenza/human_animal_interface/H5N1cumulative_table_archives/en/index.html.5. Nicholson K, Webster RG, Hay AJ. Textbook of Influenza. 1998. Blackwell Science Ltd..6. Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, et al. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med. 2001. 7:1306–1312.
Article7. Wise HM, Foeglein A, Sun J, Dalton RM, Patel S, Howard W, et al. A complicated message: Identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J Virol. 2009. 83:8021–8031.
Article8. Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, et al. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science. 2012. 337:199–204.
Article9. Inglis SC, Brown CM. Spliced and unspliced RNAs encoded by virion RNA segment 7 of influenza virus. Nucleic Acids Res. 1981. 9:2727–2740.
Article10. Lamb RA, Lai CJ, Choppin PW. Sequences of mRNAs derived from genome RNA segment 7 of influenza virus: colinear and interrupted mRNAs code for overlapping proteins. Proc Natl Acad Sci U S A. 1981. 78:4170–4174.
Article11. Lamb RA, Lai CJ. Sequence of interrupted and uninterrupted mRNAs and cloned DNA coding for the two overlapping nonstructural proteins of influenza virus. Cell. 1980. 21:475–485.
Article12. Eisinger J, Feuer B, Yamane T. Codon-anticodon binding in tRNAphe. Nat New Biol. 1971. 231:126–128.
Article13. Dias A, Bouvier D, Crépin T, McCarthy AA, Hart DJ, Baudin F, et al. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature. 2009. 458:914–918.
Article14. Hara K, Schmidt FI, Crow M, Brownlee GG. Amino acid residues in the N-terminal region of the PA subunit of influenza A virus RNA polymerase play a critical role in protein stability, endonuclease activity, cap binding, and virion RNA promoter binding. J Virol. 2006. 80:7789–7798.
Article15. Shi M, Jagger BW, Wise HM, Digard P, Holmes EC, Taubenberger JK. Evolutionary Conservation of the PA-X Open Reading Frame in Segment 3 of Influenza A Virus. J Virol. 2012. 86:12411–12413.
Article16. McAuley JL, Hornung F, Boyd KL, Smith AM, McKeon R, Bennink J, et al. Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe. 2007. 2:240–249.
Article17. Zamarin D, Ortigoza MB, Palese P. Influenza A virus PB1-F2 protein contributes to viral pathogenesis in mice. J Virol. 2006. 80:7976–7983.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Prevention and Treatment of Influenza
- Benign Acute Childhood Myositis Associated with Influenza B Virus
- Reversible Splenial Lesion in a Patient with Influenza B Virus Infection
- Atypical Kawasaki Disease Presenting as Acute Kidney Injury in a Patient with Influenza B Virus Infection
- A Case of Severe Influenza Infection in a Child with Nephrotic Syndrome on Steroid Therapy