J Bacteriol Virol.
2009 Dec;39(4):269-276. 10.4167/jbv.2009.39.4.269.
Bacteriocin from Purple Nonsulfur Phototrophic Bacteria, Rhodobacter capsulatus
- Affiliations
-
- 1Department of Life Science, Kyonggi University, Suwon, Korea. jkimtamu@kgu.ac.kr
- 2Genomictree, Inc., Daejon, Korea.
- 3School of Life & Food Sciences, Handong Global University, Pohang, Korea.
- 4Department of Biological Sciences, Sungkyunkwan University, Suwon, Korea.
- KMID: 2168538
- DOI: http://doi.org/10.4167/jbv.2009.39.4.269
Abstract
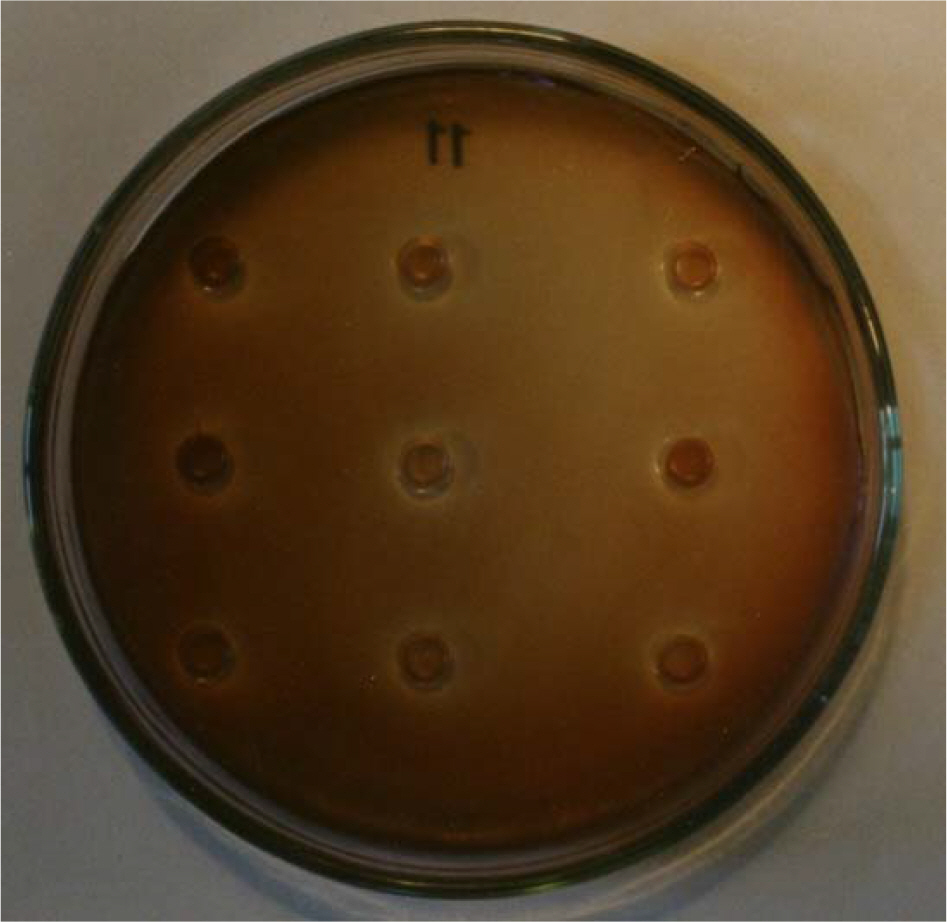
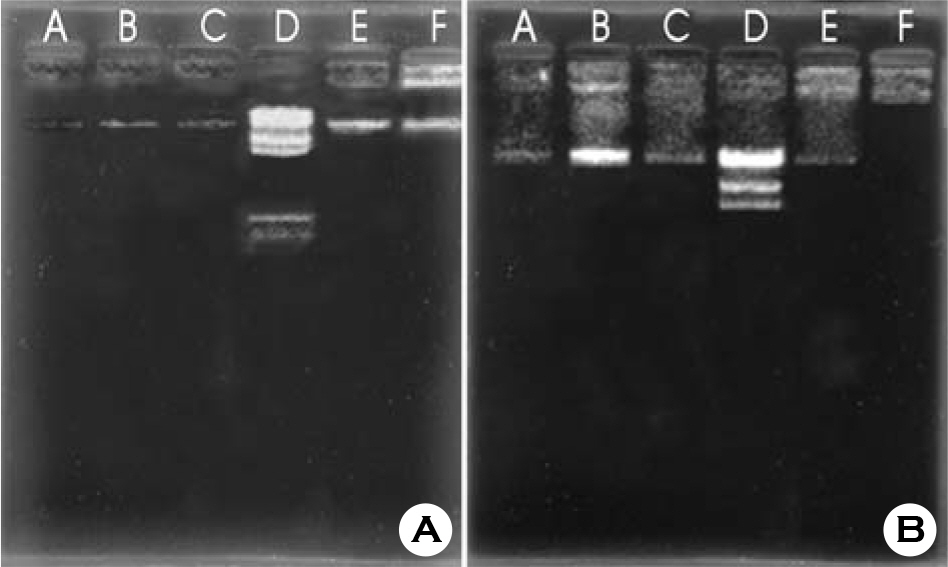
- To find whether productivity of bacteriocin is controlled between different species under unusual cultural conditions, we used Rhodobacter capsulatus ATCC 17016 as a producer and Rhodopseudomonas palustris ATCC 17003 as an indicator. Rhodobacter capsulatus was cultured under aerobic conditions in the dark in Lascelles medium containing 0.3% Triton X-100. As a result, bacteriocin productivity increased enormously. The optimal pH range of bacteriocin production was 6~7.8. Through partial purification of bacteriocin, the molecular weight was roughly estimated at 14 kDa. Plasmid had no influence on bacteriocin production by Rhodobacter capsulatus. Our findings indicate that culture conditions affect bacteriocin productivity between more distantly related species, and bacteriocin of Rhodobacter capsulatus is not encoded by a plasmid.
MeSH Terms
Figure
Reference
-
1). Brede DA., Faye T., Johnsborg O., Odegard I., Nes IF., Holo H. Molecular and genetic characterization of propionicin F, a bacteriocin from Propionibacterium freudenreichii. Appl Environ Microbiol. 2004. 70:7303–10.2). Drider D., Fimland G., Héchard Y., McMullen LM., Prévoste H. The continuing story of class IIa bacteriocins. Microbiol Mol Biol Rev. 2006. 70:564–82.
Article3). Johnsen L., Fimland G., Nissen-Meyer J. The C-terminal domain of pediocin-like antimicrobial peptides (class IIa bacteriocins) is involved in specific recognition of the C-terminal part of cognate immunity proteins and in determining the antimicrobial spectrum. J Biol Chem. 2005. 280:9243–50.
Article4). Nagao J., Asaduzzaman SM., Aso Y., Okuda K., Nakayama J., Sonomoto K. Lantibiotics: Insight and foresight for new paradigm. J Biosci Bioeng. 2006. 102:139–49.
Article5). Reeves P. The bacteriocins. USA: New York: Springer-Verlag. 1972.6). Lyon WJ., Glatz BA. Partial purification and characterization of a bacteriocin produced by Propionibacterium thoenii. Appl Environ Microbiol. 1991. 57:701–6.7). Sprules T., Kawulka KE., Vederas JC. NMR solution structure of ImB2, a protein conferring immunity to antimicrobial activity of the type IIa bacteriocin, carnobacteriocin B2. Biochemistry. 2004. 43:11740–9.
Article8). Tagg JR., Dajani AS., Wannamaker LW. Bacteriocins of gram positive bacteria. Bacteriol Rev. 1976. 40:722–56.9). O'Sullivan L., Ross RP., Hill C. Potential of bacteriocin-producing lactic acid bacteria for improvements in food safety and quality. Biochimie. 2002. 84:593–604.10). Giacometti A., Cirioni O., Barchiesi F., Fortuna M., Scalise G. In-vitro activity of cationic peptides alone and in combination with clinically used antimicrobial agents against Pseudomonas aeruginosa. J Antimicrob Chemother. 1999. 44:641–5.11). Todorov S., Gotcheva B., Dousset X., Onno B., Ivanova I. Influence of growth medium on bacteriocin production in Lactobacillus plantarum ST31. Biotechnol Biotechnol Eq. 2000. 14:50–5.12). Guest JR. Bacteriocinogeny in the Arthiorhodaceae. J Gen Microbiol. 1974. 81:513–5.13). Wall JD., Weaver PF., Gest H. Gene transfer agents, bacteriophages and bacteriocins of Rhodopseudomonas capsulata. Arch Microbiol. 1975. 105:217–24.14). Willison JC., Magnin JP., Vignais PM. Isolation and characterization of Rhodobacter capsulatus strains lacking endogenous plasmids. Arch Microbiol. 1987. 147:134–42.15). Dufour A., Hindre T., Haras D., Le Pennec JP. The biology of lantibiotics from the lacticin 481 group is coming of age. FEMS Microbiol Rev. 2007. 31:134–67.
Article16). Kawai Y., Kemperman R., Kok J., Saito T. The circular bacteriocins gassericin A and circularin A. Curr Protein Pept Sci. 2004. 5:393–8.
Article17). Maqueda M., Galvez A., Bueno MM., Sanchez-Barrena MJ., Gonzalez C., Albert A., Rico M., Valdivia E. Peptide AS-48: prototype of a new class of cyclic bacteriocins. Curr Protein Pept Sci. 2004. 5:399–416.
Article18). Lascelles J. The synthesis of porphyrins and bacteriochlorophyll by cell suspensions of Rhodopseudomonas sphaeroides. Biochem J. 1956. 62:78–93.19). Brock TD., Davie JM. Probable identity of a group D hemolysin with a bacteriocin. J Bacteriol. 1963. 86:708–12.20). Jorger MC., Klaenhammer TR. Characterization and purification helveticin J and evidence for a chromosomally determined bacteriocin produced Lactobacillus helveticus 481. J Bacteriol. 1986. 167:439–46.21). Ahn C., Stiles ME. Plasmid-associated bacteriocin production by a strain of Carnobacterium piscicola from meat. Appl Environ Microbiol. 1990. 56:2503–10.22). Hardy KG. Bacteriocins. In:. Burns RG, Slater JH, editors. editors.Experimental microbial ecology. USA St. Louis: Blackwell;1982. p. 368–78.23). Scherwitz KM., Baldwin KA., Mokay LL. Plasmid linkage of a bacteriocin-like substance in Streptococcus lactis subsp. Diacetylactis strain WM4: Transferability to Streptococcus lactis. Appl Environ Microbiol. 1983. 45:1506–12.24). Ike Y., Clewell DB., Segarra RA., Gilmore MS. Genetic analysis of the pAD1 hemolysin/bacteriocin determinant in Enterococcus faecalis: Tn917 insertional mutagenesis and cloning. J Bacteriol. 1990. 172:155–63.25). Yule R., Barridge BD. Isolation and characterization of a bacteriocin produced by Bacillus stearothermophilus strain NU-10. Can J Microbiol. 1976. 22:1743–50.26). Maniatis T., Fritsch EF., Sambrook J. Molecular cloning, a laboratory manual. NY: Cold Spring Harbor: Cold Spring harbor laboratory. 1982.27). Bearden JC Jr. Electrophoretic mobility of high-molecular-weight, double-stranded DNA on agarose gels. Gene. 1979. 6:221–34.
Article28). Kado CI., Liu ST. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981. 145:1365–73.
Article29). Rheinwald JG., Chakrabarty AM., Gunsalus IC. A transmissible plasmid controlling camphor oxidation in Pseudomonas putida. Proc Natl Acad Sci USA. 1973. 70:885–9.30). Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem. 1976. 72:248–54.31). Farber E. Cellular biochemistry of the stepwise development of cancer with chemicals: G. H. A. Clowes memorial lecture. Cancer Res. 1984. 44:5463–74.32). Todorov SD., Dicks LM. Influence of growth conditions on the production of a bacteriocin by Lactococcus lactis subsp. lactis ST34BR, a strain isolated from barley beer. J Basic Microbiol. 2004. 44:305–16.33). Garnier T., Cole ST. Characterization of a bacteriocinogenic plasmid from Clostridium perfringens and molecular genetic analysis of the bacteriocin-encoding gene. J Bacteriol. 1986. 168:1189–96.34). Rammelsberg M., Muller E., Radler F. Casein 80: purification characterization of a new bacteriocin from Lactobacillus casei. Arch Microbiol. 1990. 154:249–52.35). Cooper PC., Hawkins FK., James R. Incompatibility between E colicin plasmids. J Gen Microbiol. 1986. 132:1859–62.
Article36). Herschman HR., Helinski DR. Purification and characterization of colicin E2 and colicin E3. J Biol Chem. 1967. 242:5360–8.37). Southern JA., Katz W., Woods DR. Purification and properties of a cell-bound bacteriocin from a Bacteroides fragilis strain. Antimicrob Agents Chemother. 1984. 25:253–7.38). Austin-Prather SL., Booth SJ. Evidence for a membrane-bound form of a bacteriocin of Bacteroides uniformis Tl-1. Can J Microbiol. 1984. 30:268–72.