J Bacteriol Virol.
2008 Mar;38(1):1-10. 10.4167/jbv.2008.38.1.1.
Roles of Flagellar Hook-Associated Proteins in Vibrio vulnificus Motility and Virulence
- Affiliations
-
- 1Clinical Vaccine R&D Center, Chonnam National University Hwasun Hospital, Jeonnam, Korea. jhrhee@chonnam.ac.kr
- 2Research Institute of Vibrio Infections and Genome Research Center for Enteropathogenic Bacteria, Chonnam National University Medical School, Gwangju, Korea.
- 3National Research Laboratory of Molecular Microbial Pathogenesis and Department of Microbiology, Chonnam National University Medical School, Gwangju, Korea.
- KMID: 2168513
- DOI: http://doi.org/10.4167/jbv.2008.38.1.1
Abstract
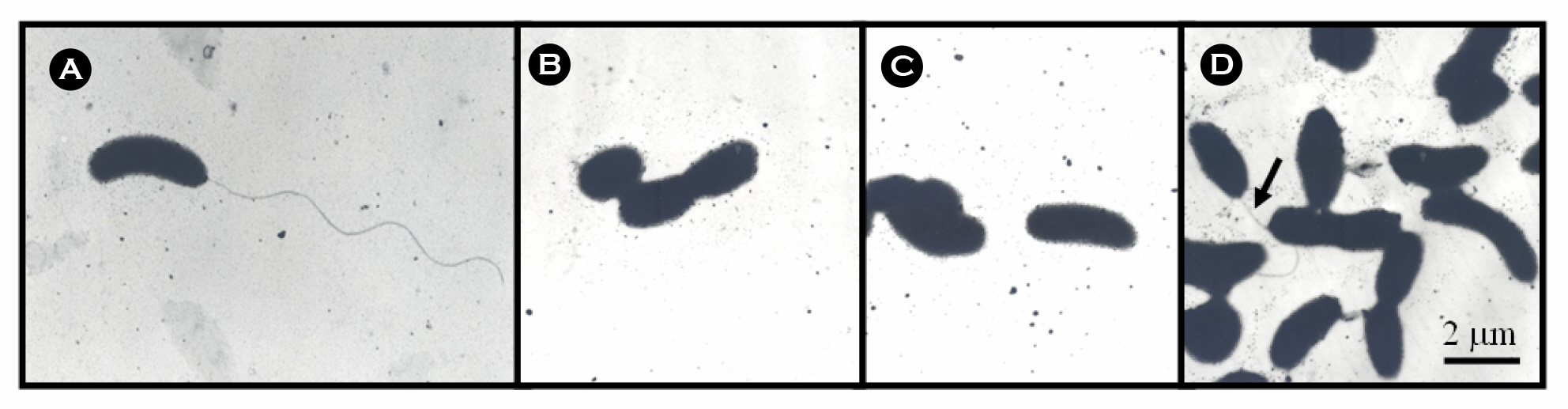
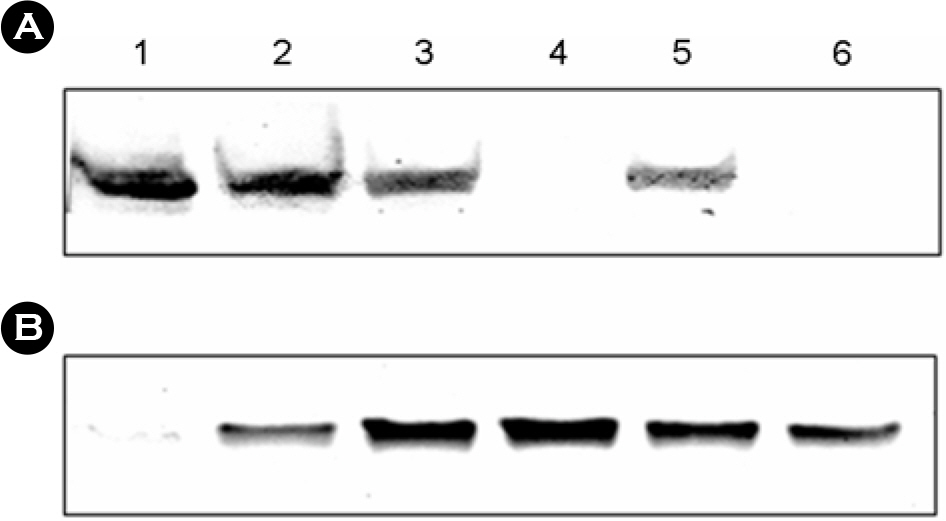
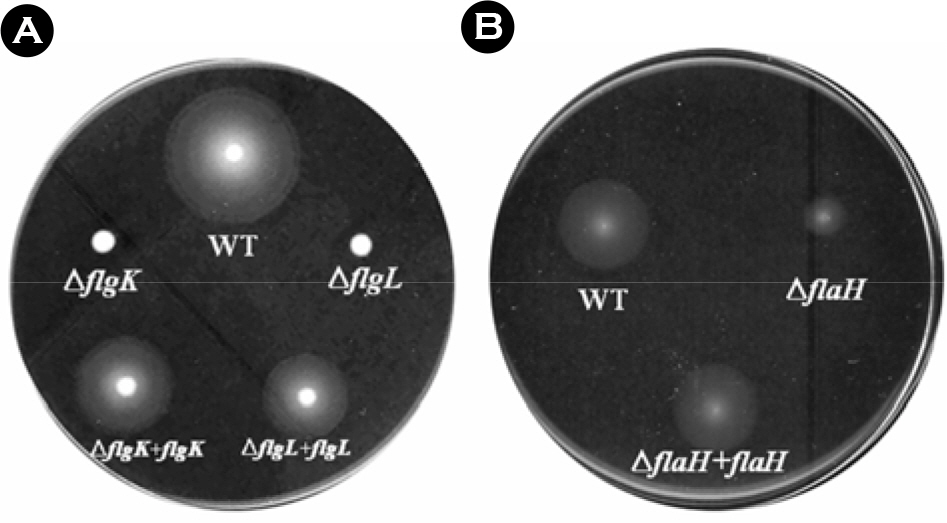
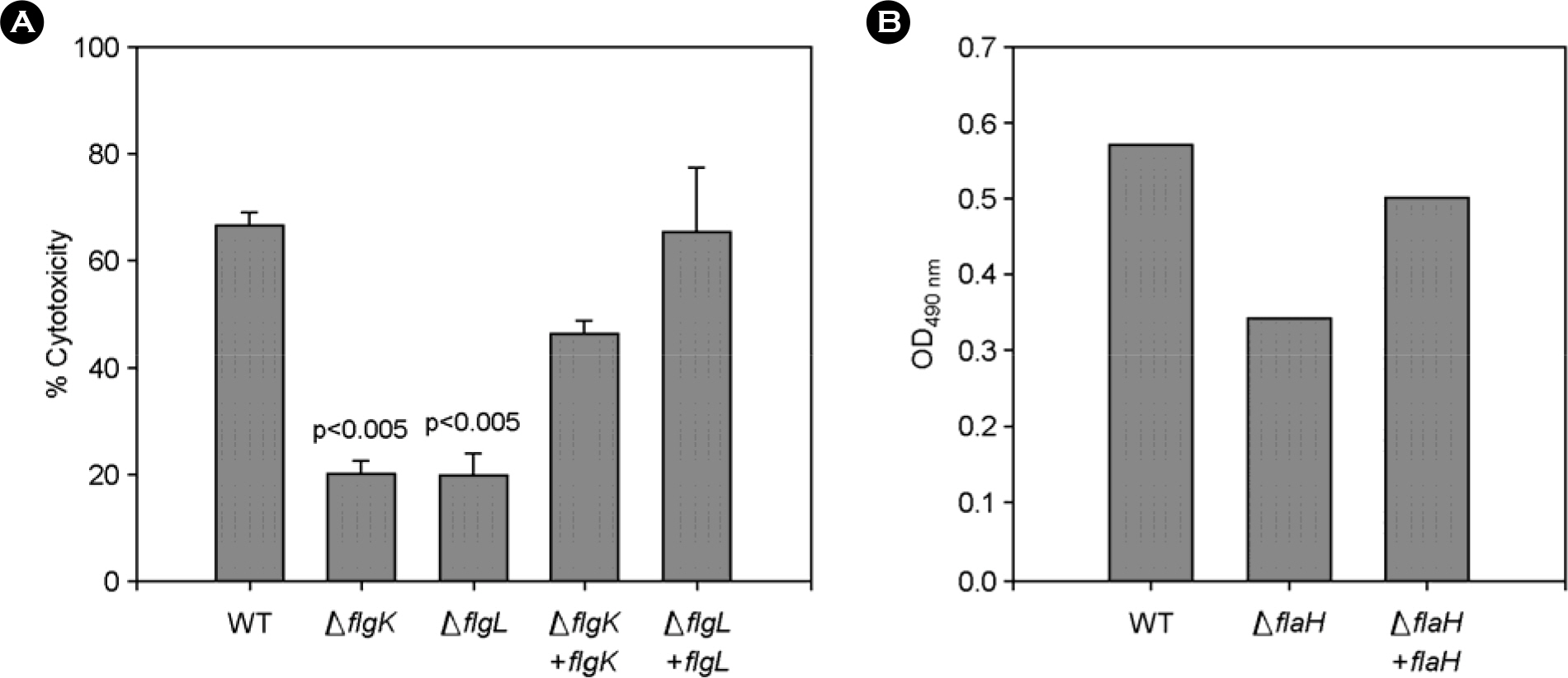
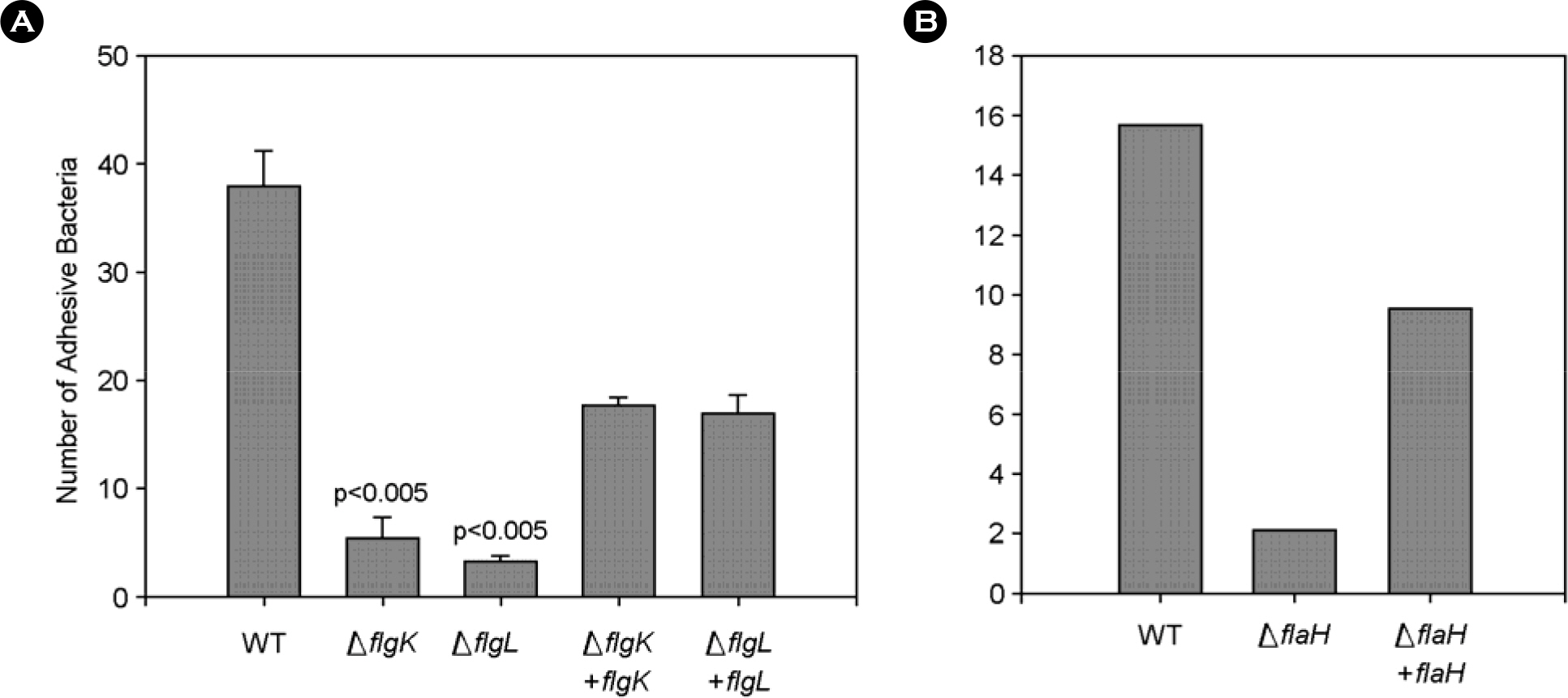
- The bacterial flagellar structure can be divided into the basal body, the hook and the filament. Three minor components called hook associated proteins (HAP1, HAP2 and HAP3) form a junction between the hook and the filament (HAP1 and HAP3) and a capping structure at the distal end of flagellar filament (HAP2). Vibrio vulnificus is a halophilic pathogenic bacterium that is locomotive by means of a polar flagellum. From a V. vulnificus genome sequencing project, we obtained sequences of V. vulnificus flgK (Vv-flgK), flgL (Vv-flgL), and flaH (Vv-flaH) genes that encode HAP1, HAP3, and HAP2, respectively. To investigate roles of the HAP proteins, deletion mutants of the Vv-flgK, Vv-flgL and Vv-flaH were constructed. Electron microscopic analysis showed that the Vv-flgK or Vv-flgL mutant did not produce an intact polar flagellum while the Vv-flaH mutant produced a fragile flagellar structure. Western blot analysis against a major polar flagellin proposed that the null HAP1 and HAP3 mutations resulted in a failure of normal flagellar assembly since flagellins produced by the mutants were secreted out in the culture supernatants without long flagellar filaments. Motility was completely abolished by a single mutation in HAP1 or HAP3, and the HAP2 mutant showed a decreased motility. Also each of the mutants showed an impaired cytotoxicity and adherence to HeLa cell compared with the isogenic wild type strain. LD(50) increased by 10- and 11-fold in the V. vulnificus HAP3 and HAP2 mutant, respectively. These results suggest that the HAP proteins play important roles in polar flagellation and the virulence of V. vulnificus.
MeSH Terms
Figure
Reference
-
References
1). Ditta G, Stanfield S, Corbin D, Helinski DR. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 77:7347–7351. 1980.2). Furukawa Y, Imada K, Vonderviszt F, Matsunami H, Sano K, Kutsukake K, Namba K. Interactions between bacterial flagellar axial proteins in their monomeric state in solution. J Mol Biol. 318:889–900. 2002.
Article3). Girón JA, Torres AG, Freer E, Kaper JB. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol Microbiol. 44:361–379. 2002.4). Gray LD, Kreger AS. Purification and characterization of an extracellular cytolysin produced by Vibrio vulnificus. Infect Immun. 48:67–72. 1985.5). Hayat U, Reddy GP, Bush CA, Johnson JA, Wright AC, Morris JG Jr. Capsular types of Vibrio vulnificus: an analysis of strains from clinical and environmental sources. J Infect Dis. 168:758–762. 1993.6). Homma M, Fujita H, Yamaguchi S, Iino T. Excretion of unassembled flagellin by Salmonella typhimurium mutants deficient in hook-associated proteins. J Bacteriol. 159:1056–1059. 1984.7). Homma M, Iino T. Excretion of unassembled hook-associated proteins by Salmonella typhimurium. J Bacteriol. 164:1370–1372. 1985.8). Homma M, Iino T. Locations of hook-associated proteins in flagellar structures of Salmonella typhimurium. J Bacteriol. 162:183–189. 1985.9). Homma M, Kutsukake K, Iino T. Structural genes for flagellar hook-associated proteins in Salmonella typhimurium. J Bacteriol. 163:464–471. 1985.10). Homma M, Kutsukake K, Iino T, Yamaguchi S. Hook-associated proteins essential for flagellar filament formation in Salmonella typhimurium. J Bacteriol. 157:100–108. 1984.11). Ikeda T, Asakura S, Kamiya R. “Cap” on the tip of Salmonella flagella. J Mol Biol. 184:735–737. 1985.12). Ikeda T, Homma M, Iino T, Asakura S, Kamiya R. Localization and stoichiometry of hook-associated proteins within Salmonella typhimurium flagella. J Bacteriol. 169:1168–1173. 1987.13). Ikeda T, Yamaguchi S, Hotani H. Flagellar growth in a filament-less Salmonella fliD mutant supplemented with purified hook-associated protein 2. J Biochem. 114:39–44. 1993.14). Kim SY, Kim YR, Hong HY, Ryu PY, Choy HE, Chung SS, Rhee JH. Polar flagellin FlaB of Vibrio vulnificus serves an important virulence factor for adhesion and invasion. In American Society for Microbiology General Meeting. Washington, DC: American Society for Microbiology Press, abstract. B-145:pp.56. 2002.15). Kim SY, Lee SE, Kim YR, Kim CM, Ryu PY, Choy HE, Chung SS, Rhee JH. Regulation of Vibrio vulnificus virulence by the LuxS quorum-sensing system. Mol Microbiol. 48:1647–1664. 2003.16). Kim YK, McCarter LL. Analysis of the polar flagellar gene system of Vibrio parahaemolyticus. J Bacteriol. 182:3693–3704. 2000.17). Kim YR, Kim CM, Rhee JH. Close encounter of Vibrio vulnificus with host cells is a prerequisite to the cytotoxicity of yet unidentified new virulence factor. J Bacteriol and Virol. 33:19–26. 2003.18). Kim YR, Lee SE, Kook H, Yeom JA, Na HS, Kim SY, Chung SS, Choy HE, Rhee JH. Vibrio vulnificus RTX toxin kills host cells only after contact of the bacteria with host cells. Cell Microbiol. 10:848–862. 2008.19). Kothary MH, Kreger AS. Purification and characterization of an elastolytic protease of Vibrio vulnificus. J Gen Microbiol. 133:1783–1791. 1987.20). Litwin CM, Rayback TW, Skinner J. Role of catechol siderophore synthesis in Vibrio vulnificus virulence. Infect Immun. 64:2834–2838. 1996.21). McCarter LL. Genetic and molecular characterization of the polar flagellum of Vibrio parahaemolyticus. J Bacteriol. 177:1595–1609. 1995.22). McCarter LL. Polar flagellar motility of the Vibrionaceae. Microbiol Mol Biol Rev. 65:445–462. 2001.23). Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 170:2575–2583. 1988.24). Milton DL, O'Toole R, Horstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J Bacteriol. 178(5):1310–1319. 1996.25). Morris JG Jr, Wright AC, Roberts DM, Wood PK, Simpson LM, Oliver JD. Identification of environmental Vibrio vulnificus isolates with a DNA probe for the cytotoxin-hemolysin gene. Appl Envion Microbiol. 53:193–195. 1987.26). Paik KW, Moon B, Park CW, Kim KY, Ji MS, Choi SK, Rew JS, Yoon CM. Clinical characteristics of ninety-two cases of Vibrio vulnificus infections. Kor J Infect Dis. 27:355–365. 1995.27). Park SD, Shon HS, Joh NJ. Vibrio vulnificus septicemia in Korea: clinical and epidemiologic findings in seventy patients. J Am Acad Dermatol. 24:397–403. 1991.28). Reed LJ, Muench H. A simple method of estimating the fifty percent endpoints. Am J Hyg. 27:493–497. 1938.29). Reyrat JM, Pelicic V, Gicquel B, Rappuoli R. Counter-selectable markers: Untapped tools for bacterial genetics and pathogenesis. Infect Immun. 66:4011–4017. 1998.
Article30). Simon R, Prifer R, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1:784–791. 1983.31). Tacket CO, Brenner F, Blake PA. Clinical features and epidemiological study of Vibrio vulnificus infections. J Infect Dis. 149:558–561. 1984.32). Wright AC, Morris JG Jr. The extracellular cytolysin of Vibrio vulnificus: Inactivation and relationship to virulence in mice. Infect Immun. 59:192–197. 1991.33). Yonekura K, Maki-Yonekura S, Namba K. Structure analysis of the flagellar cap-filament complex by electron cryomicroscopy and single-particle image analysis. J Struct Biol. 133:246–253. 2001.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Quorum Sensing System and Virulence Regulation in Vibrio vulnificus
- Effect of Hydrogen Ion Concentration on the Growth of Vibrio vulnificus
- Multiple High-affinity Iron-uptake Systems of Vibrio vulnificus
- A Case of Acute Gastroenteritis caused by Vibrio parahaemolyticus and Vibrio vulnificus
- Production of and sensitivity to bacteriocin, "vulnificin" of vibrio vulnificus