J Bacteriol Virol.
2015 Jun;45(2):132-137. 10.4167/jbv.2015.45.2.132.
Molecular Epidemiology of Mumps Virus Circulated in Gwangju
- Affiliations
-
- 1Health and Environment Institute of Gwangju, Gwangju, Korea. jkchung@korea.kr
- 2National Institute of Health, Korea Centers for Disease Control and Prevection, Chungcheongbuk-do, Korea.
- KMID: 2149213
- DOI: http://doi.org/10.4167/jbv.2015.45.2.132
Abstract
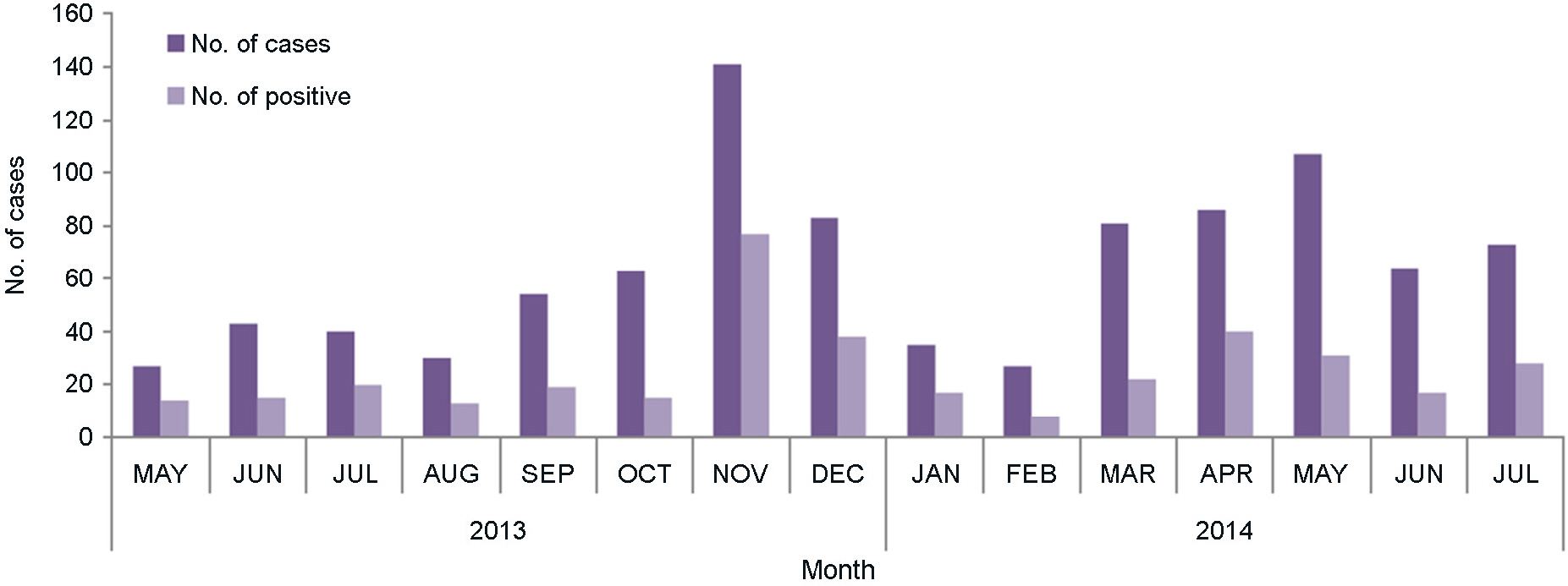
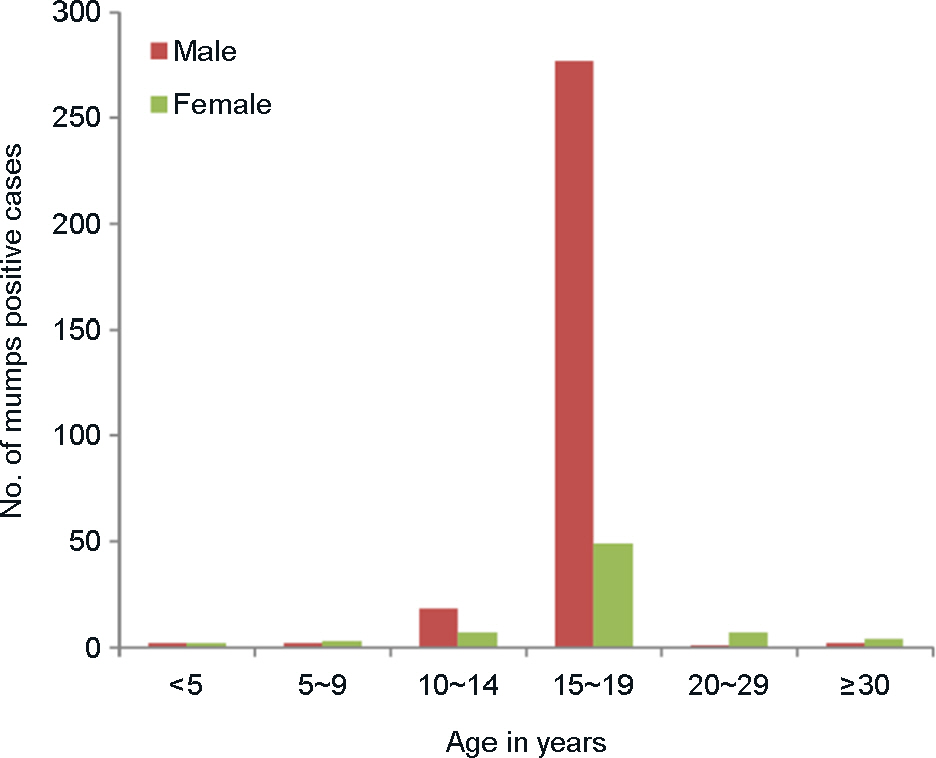
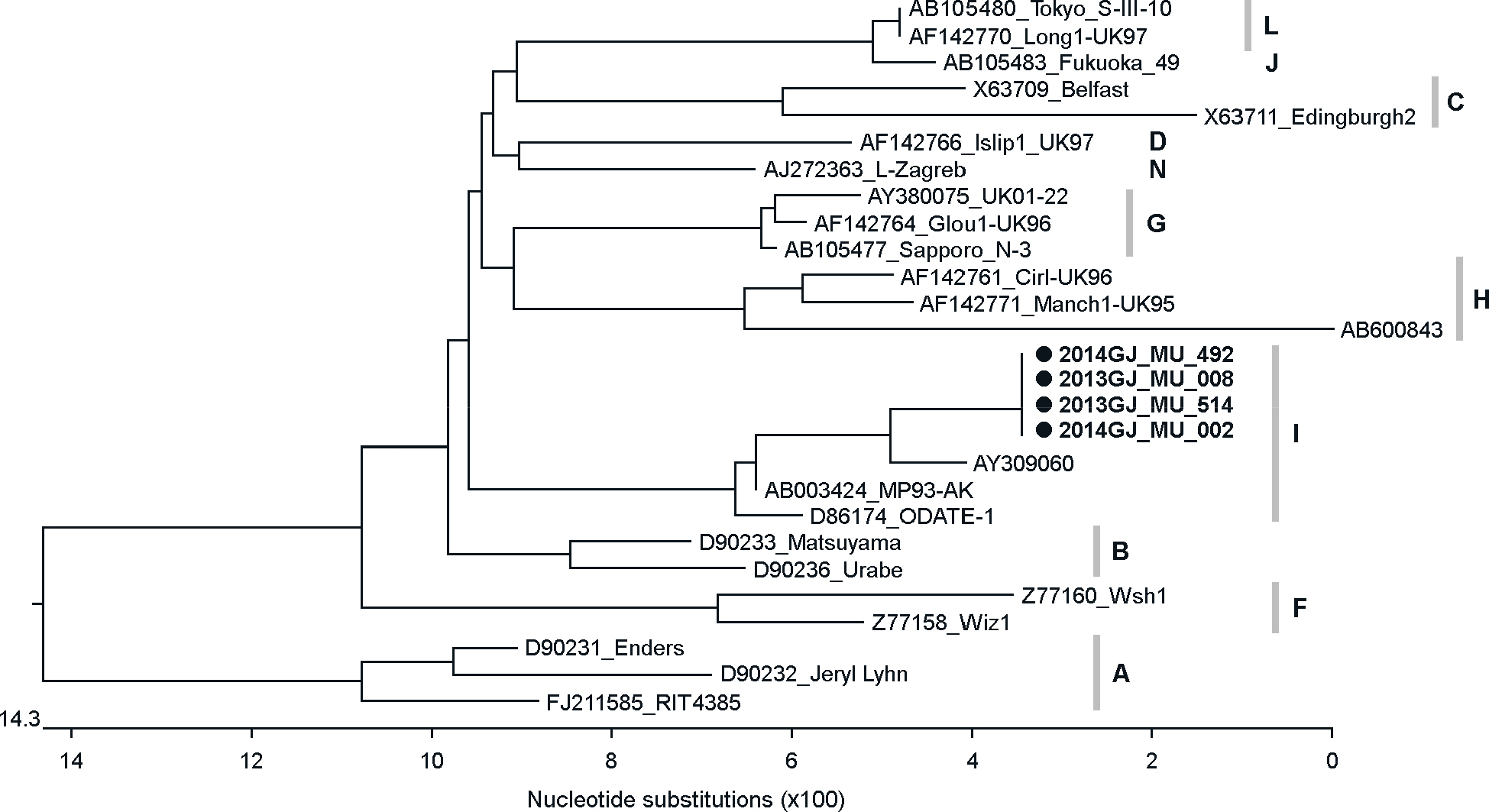
- The recent mumps epidemic in South Korea has generated a large amount of public concern. This study has attempted to analyze molecular epidemiological changes of mumps virus circulating in Gwangju metropolitan area, South Korea. 953 throat swab samples were collected from patients with parotitis from May 2013 to July 2014. The majority (71.5%) of these cases have occurred in middle or high school students aged from 15 to 19 years. All samples were tested using a reverse transcription polymerase chain reaction (RT-PCR) that targets the short hydrophobic (SH) gene of the virus. Mumps virus SH gene was detected in 39.2% (374/953) of samples. And 82 RT-PCR products were randomly selected for nucleotide sequencing analysis. All of these sequences were determined as genotype I by phylogenetic analysis and showed the highest nucleic acid similarity (99%) with Dg1062/Korea/98 (GenBank accession no. AY309060). These results suggested that appearance of new genotype or genetic variation at the nucleotide level could be ruled out to evaluate main cause of recent mumps outbreak in Gwangju metropolitan area.
Keyword
MeSH Terms
Figure
Reference
-
1). Steven AR, Christian JS, Kathryn MC. Mumps Virus. Davis MK, Peter MH, editors. Fields Virology. 6th ed.Philadelphia: Lippincott Williams & Wilkins;2013. p. 1024–41.2). Jin L, Örvell C, Myers R, Rota PA, Nakayama T, Forcic D, et al. Genomic diversity of mumps virus and global distribution of the 12 genotypes. Rev Med Virol. 2015; 25:85–101.
Article3). Korea Centers for Disease Control and Prevention. Disease web statistics system. Available from:. http://is.cdc.go.kr/nstat/index.jsp.4). Barskey AE, Glasser JW, LeBaron CW. Mumps resurgences in the United States: A historical perspective on unexpected elements. Vaccine. 2009; 27:6186–95.
Article5). Kaaijk P, Van der Zeijst B, Boog M, Hoitink C. Increased mumps incidence in the Netherlands: review on the possible role of vaccine strain and genotype. Euro Surveill. 2008; 13.
Article6). Sane J, Gouma S, Koopmans M, de Melker H, Swaan C, van Binnendijk R, et al. Epidemic of mumps among vaccinated persons, the Netherlands, 2009–2012. Emerg Infect Dis. 2014; 20:643–8.
Article7). Park DW, Nam MH, Kim JY, Kim HJ, Sohn JW, Cho Y, et al. Mumps outbreak in a highly vaccinated school population: Assessment of secondary vaccine failure using IgG avidity measurements. Vaccine. 2007; 25:4665–70.
Article8). Ryu JU, Kim EK, Youn YS, Rhim JW, Lee KY. Outbreaks of mumps: an observational study over two decades in a single hospital in Korea. Korean J Pediatr. 2014; 57:396–402.
Article9). Okajima K, Iseki K, Koyano S, Kato A, Azuma H. Virological analysis of a regional mumps outbreak in the northern island of Japan-mumps virus genotyping and clinical description. Jpn J Infect Dis. 2013; 66:561–3.10). Davidkin I, Jokinen S, Paananen A, Leinikki P, Peltola H. Etiology of mumps-like illnesses in children and adolescents vaccinated for measles, mumps, and rubella. J Infect Dis. 2005; 191:719–23.
Article11). Barskey AE1, Juieng P, Whitaker BL, Erdman DD, Oberste MS, Chern SW, et al. Viruses detected among sporadic cases of parotitis, United States, 2009–2011. J Infect Dis. 2013; 208:1979–86.
Article12). Palacios G, Jabado O, Cisterna D, de Ory F, Renwick N, Echevarria JE, et al. Molecular identification of mumps virus genotypes from clinical samples: standardized method of analysis. J Clin Microbiol. 2005; 43:1869–78.
Article13). Lee JY, Kim YY, Shin GC, Na BK, Lee JS, Lee HD, et al. Molecular characterization of two genotypes of mumps virus circulated in Korea during 1998–2001. Virus Res. 2003; 97:111–6.14). Lee JY, Na BK, Kim JH, Lee JS, Park JW, Shin GC, et al. Regional outbreak of mumps due to genotype H in Korea in 1999. J Med Virol. 2004; 73:85–90.
Article15). Lee JY, Na BK, Lee HD, Chang SW, Kim KA, Kim JH, et al. Complete nucleotide sequence of a mumps virus Genotype I strain isolated in Korea. Virus Genes. 2004; 28:201–5.
Article16). Kashiwagi Y, Takami T, Mori T, Nakayama T. Sequence analysis of F, SH, and HN genes among mumps virus strains in Japan. Arch Virol. 1999; 144:593–9.
Article17). Vandermeulen C, Roelants M, Vermoere M, Roseeuw K, Goubau P, Hoppenbrouwers K. Outbreak of mumps in a vaccinated child population: a question of vaccine failure? Vaccine. 2004; 22:2713–6.
Article18). Davidkin I, Jokinen S, Broman M, Leinikki P, Peltola H. Persistence of measles, mumps, and rubella antibodies in an MMR-vaccinated cohort: a 20-year follow-up. J Infect Dis. 2008; 197:950–6.