Tuberc Respir Dis.
2007 Aug;63(2):128-138. 10.4046/trd.2007.63.2.128.
Mutations of katG and inhA in MDR M. tuberculosis
- Affiliations
-
- 1Department of Microbiology, Seoul National University College of Medicine, Seoul, Korea. yhkook@sun.ac.kr
- 2Cancer Research Institute, Seoul National University College of Medicine, Seoul, Korea.
- 3Institute of Endemic Diseases, SNUMRC, Seoul National University College of Medicine, Seoul, Korea.
- 4Clinical Research Institute, Seoul National University Hospital, Seoul, Korea.
- 5The Korean Institute of Tuberculosis, The Korean National Tuberculosis Association, Korea.
- KMID: 1910091
- DOI: http://doi.org/10.4046/trd.2007.63.2.128
Abstract
-
BACKGROUNDS: Mutations of katG and inhA (ORF and promoter) are known to be related to isoniazid (INH) resistance of Mycobacterium tuberculosis. Because reports on these mutations in Korean isolates are limited (i.e. only the frequency of katG codon 463 was evaluated.), we tried to know the kinds of mutations of two genes and their frequencies in INH resistant Korean M. tuberculosis strains.
METHODS
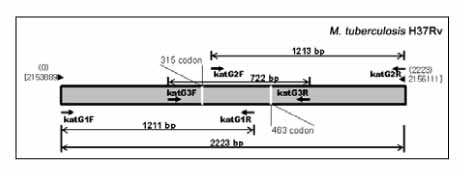
PCR was performed to amplify katG (2,223 bp), inhA ORF (-77~897, 975 bp), and inhA promoter (-168~80, 248 bp) from 29 multidrug resistant M. tuberculosis (MDR-TB) DNAs prepared by bead beater-phenol method. Their sequences were determined and analyzed by ABI PRISM 3730 XL Analyzer and MegAlign package program, respectively.
RESULTS
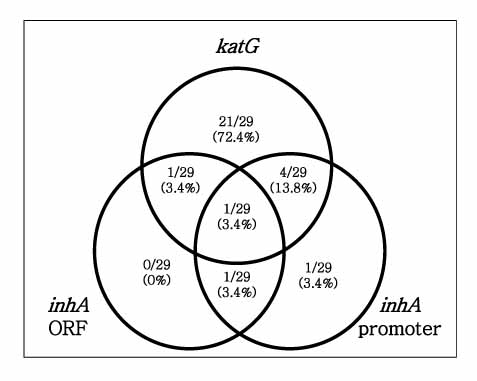
All of the isolates had more than one mutation in katG or inhA gene. Twenty seven (93%) of 29 tested strains had katG mutations, which suggests that katG is a critical gene determining INH resistance of M. tuberculosis. Amino acid substitutions, such as Arg463Leu and Ser315Thr, due to point mutations of the katG were the most frequent (62.1% and 55.2%) mutations. In addition, deletion of the katG gene was frequently observed (17.2%). Analyzed Korean MDR-TB isolates also had variable inhA mutations. Point mutation of inhA promoter region, such as -15 (C-->T) was frequently found. Substitution of amino acid (Lsy8Asn) due to point mutation (AAA-->AAC) of inhA ORF was found in 1 isolate. Interestingly, 14 point mutated types that were not previously reported were newly found. While four types resulted in amino acid change, the others were silent mutations.
CONCLUSIONS
Although it is not clear that the relationship of these newly found mutations with INH resistance, they show marked diversity in Korean MDR-TB strains. It also suggests their feasibility as a molecular target to supplement determining the INH resistance of clinical isolates because of the possible existence of low-level INH resistant strains.
MeSH Terms
Figure
Reference
-
1. Dye C, Scheele S, Dolin P, Pathania V, Raviglione MC. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA. 1999. 282:677–686.2. Bai GH. Anti-tuberculosis drug resistance in Korea. Communicable Diseases Monthly Report. 2005. 16:101–107.3. Crofton J, Chaulet P, Maher D, Grosset J, Harris W, Norman H, et al. Guidelines for the management of drug-resistant tuberculosis. 1997. Geneva: World Health Organization.4. World Health Organization. Anti-tuberculosis drug resistance in the world: the WHO/IUATLD global project on antituberculosis drug resistance surveillance 1994-1997. 1997. Geneva: World Health Organization.5. Kim SY, Lee JY, Ryu SR, Kim SJ, Bai GH. Rapid diagnosis of isoniazid resistance by detection of mutations in katG and inhA of Mycobacterium tuberculosis in Korea. J Bacteriol Virol. 1997. 32:569–576.6. Park YK, Shim MS, Cho SH, Bai GH, Kim SJ. The relationship between isoniazid resistance and 463 codon mutation of katG gene in Mycobacterium tuberculosis. Tuberc Respir Dis. 1996. 43:8–13.7. Dye C, Espinal MA, Watt CJ, Mbiaga C, Williams BG. Worldwide incidence of multidrug-resistant tuberculosis. J Infect Dis. 2002. 185:1197–1202.8. Morlock GP, Metchock B, Sikes D, Crawford JT, Cooksey RC. ethA, inhA, and katG loci of ethionamide-resistant clinical Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother. 2003. 47:3799–3805.9. Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase- peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992. 358:591–593.10. Heym B, Zhang Y, Poulet S, Young D, Cole ST. Characterization of the katG gene encoding a catalase-peroxidase required for the isoniazid susceptibility of Mycobacterium tuberculosis. J Bacteriol. 1993. 175:4255–4259.11. O'Brien KL, Dietz HC, Romagnoli M, Eiden J. Evaluation of inhA gene and catalase-peroxidase gene among isoniazid-sensitive and resistant Mycobacterium tuberculosis isolates. Mol Cell Probes. 1996. 10:1–6.12. Lee AS, Tang LL, Lim IH, Ling ML, Tay L, Wong SY. Lack of clinical significance for the common arginine-to-leucine substitution at codon 463 of the katG gene in isoniazid-resistant Mycobacterium tuberculosis in Singapore. J Infect Dis. 1997. 176:1125–1127.13. Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um KS, Wilson T, et al. InhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994. 263:227–230.14. Larsen MH, Vilcheze C, Kremer L, Besra GS, Parsons L, Salfinger M, et al. Overexpression of inhA, but not kasA, confers resistance to isoniazid and ethionamide in Mycobacterium smegmatis, M. bovis BCG and M. tuberculosis. Mol Microbiol. 2002. 46:453–466.15. Basso LA, Zheng R, Musser JM, Jacobs WR, Blanchard JS. Mechanisms of isoniazid resistance in Mycobacterium tuberculosis: enzymatic characterization of enoyl reductase mutants identified in isoniazid resistant clinical isolates. J Infect Dis. 1998. 178:769–775.16. Kapur V, Li LL, Hamrick MR, Plikaytis BB, Shinnick TM, Telenti A, et al. Rapid Mycobacterium species assignment and unambiguous identification of mutations associated with antimicrobial resistance in Mycobacterium tuberculosis by automated DNA sequencing. Arch Pathol Lab Med. 1995. 119:131–138.17. Ristow M, Möhlig M, Rifai M, Schatz H, Feldmann K, Pfeiffer A. New isoniazid/ethionamide resistance gene mutation and screening for multidrug-resistant Mycobacterium tuberculosis strains. Lancet. 1995. 346:502–503.18. Lee H, Cho SN, Bang HE, Lee JH, Bai GH, Kim SJ, et al. Exclusive mutations related to isoniazid and ethionamide resistance among Mycobacterium tuberculosis isolates from Korea. Int J Lung Dis. 2000. 4:441–447.19. Morris S, Bai GH, Suffys P, Portillo-Gomez L, Fairchok M, Rouse D. Molecular mechanisms of multiple drug resistance in clinical isolates of Mycobacterium tuberculosis. J Infect Dis. 1995. 171:954–960.20. Musser JM, Kapur V, Williams DL, Kreiswirth BN, van Soolingen D, van Embden JD. Characterization of the catalase-peroxidase gene (katG) and inhA locus in isoniazid-resistant and isoniazid-susceptible strains of Mycobacterium tuberculosis by automated DNA sequencing: restricted array of mutations associated with drug resistance. J Infect Dis. 1996. 173:196–202.21. Rouse DA, Li Z, Bai GH, Morris SL. Characterization of the katG and inhA genes of isoniazid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1995. 39:2472–2477.22. Gonzalez N, Torres MJ, Aznar J, Palomares JC. Molecular analysis of rifampin and isoniazid resistance of Mycobacterium tuberculosis clinical isolates in Seville, Spain. Tuber Lung Dis. 1999. 79:187–190.23. Heym B, Honore N, Truffot-Pernot C, Banerjee A, Schurra C, Jacobs WR, et al. Implications of multi-drug resistance for the future short-course chemotherapy of tuberculosis: a molecular study. Lancet. 1994. 344:293–298.24. Rinder H, Thomschke A, Rüsch-Gerdes S, Bretzel G, Feldmann K, Rifai M, et al. Significance of ahpC promoter mutations for the prediction of isoniazid resistance in Mycobacterium tuberculosis. Eur J Clin Microbiol Infect Dis. 1998. 17:508–511.25. Silva MS, Senna SG, Ribeiro MO, Valim AR, Telles MA, Kritski A, et al. Mutations in katG, inhA, and ahpC genes of Brazilian isoniazid-resistant isolates of Mycobacterium tuberculosis. J Clin Microbiol. 2003. 41:4471–4474.26. Lee AS, Lim IH, Tang LL, Telenti A, Wong SY. Contribution of kasA analysis to detection of isoniazid-resistant Mycobacterium tuberculosis in Singapore. Antimicrob Agents Chemother. 1999. 43:2087–2089.27. Lee AS, Teo AS, Wong SY. Novel mutations in ndh in isoniazid-resistant Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother. 2001. 45:2157–2159.28. Kim BJ, Lee SH, Lyu MA, Kim SJ, Bai GH, Chae GT, et al. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB). J Clin Microbiol. 1999. 37:1714–1720.29. Shim TS, Yoo CG, Han SK, Shim YS, Kim YW. Isoniazid resistance and the point mutation of codon 463 of katG gene of Mycobacterium tuberculosis. J Korean Med Sci. 1997. 12:92–98.30. Marttila HJ, Soini H, Huovinen P, Viljanen MK. katG mutations in isoniazid-resistant Mycobacterium tuberculosis isolates recovered from Finnish patients. Antimicrob Agents Chemother. 1996. 40:2187–2189.31. Altamirano M, Marostenmaki J, Wong A, FitzGerald M, Black WA, Smith JA. Mutations in the catalase-peroxidase gene from isoniazid resistant Mycobacterium tuberculosis isolates. J Infect Dis. 1994. 169:1162–1165.32. Cockerill FR 3rd, Uhl JR, Temesgen Z, Zhang Y, Stockman L, Roberts GD, et al. Rapid identification of a point mutation of the Mycobacterium tuberculosis catalase-peroxidase (katG) gene associated with isoniazid resistance. J Infect Dis. 1995. 171:240–245.33. Heym B, Alzari PM, Honore N, Cole ST. Missense mutations in the catalase-peroxidase gene, katG, are associated with isoniazid resistance in Mycobacterium tuberculosis. Mol Microbiol. 1995. 15:235–245.34. Rouse DA, Morris SL. Molecular mechanisms of isoniazid resistance in Mycobacterium tuberculosis and Mycobacterium bovis. Infect Immun. 1995. 63:1427–1433.35. Bakonyte D, Baranauskaite A, Cicenaite J, Sosnovskaja A, Stakenas P. Molecular characterization of isoniazid-resistant Mycobacterium tuberculosis clinical isolates in Lithuania. Antimicrob Agents Chemother. 2003. 47:2009–2011.36. Torres MJ, Criado A, Gónzalez N, Palomares JC, Aznar J. Rifampin and isoniazid resistance associated mutations in Mycobacterium tuberculosis clinical isolates in Seville, Spain. Int J Tuberc Lung Dis. 2002. 6:160–163.37. Pretorius GS, van Helden PD, Sirgel F, Eisenach KD, Victor TC. Mutations in katG gene sequences in isoniazid-resistant clinical isolates of Mycobacterium tuberculosis are rare. Antimicrob Agents Chemother. 1995. 39:2276–2281.38. Bobadilla-del-Valle M, Ponce-de-Leon A, Arenas-Huertero C, Vargas-Alarcon G, Kato-Maeda M, Small PM, et al. rpoB gene mutations in rifampin-resistant Mycobacterium tuberculosis identified by polymerase chain reaction single-stranded conformational polymorphism. Emerg Infect Dis. 2001. 7:1010–1013.39. Moghazeh SL, Pan X, Arain T, Stover CK, Musser JM, Kreiswirth BN. Comparative antimycobacterial activities of rifampin, rifapentine, and KRM-1648 against a collection of rifampin-resistant Mycobacterium tuberculosis isolates with known rpoB mutations. Antimicrob Agents Chemother. 1996. 40:2655–2657.40. Williams DL, Spring L, Collins L, Miller LP, Heifets LB, Gangadharam PR, et al. Contribution of rpoB mutations to development of rifamycin cross-resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1998. 42:1853–1857.41. Bodmer T, Zürcher G, Imboden P, Telenti A. Mutation position and type of substitution in the β-subunit of the RNA polymerase influence in-vitro activity of rifamycins in rifampicin-resistant Mycobacterium tuberculosis. J Antimicrob Chemother. 1995. 35:345–348.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Correlation between GenoType MTBDRplus Assay and Phenotypic Susceptibility Test for Prothionamide in Patients with Genotypic Isoniazid Resistance
- Rapid Drug Susceptibility Testing for Isoniazid and Rifampicin by Reverse Hybridization Assay
- Molecular analysis of katG gene mutations in strains of Mycobacterium tuberculosis from Korea
- Evaluation of the AdvanSure MDR-TB GenoBlot Assay for Detection of Rifampin and Isoniazid Resistant Mycobacterium tuberculosis Complex in Respiratory Specimens
- Rapid Diagnosis of Isoniazid Resistance by Detection of Mutations in katG and inhA of Mycobacterium tuberculosis from Korea