Brain Tumor Res Treat.
2015 Apr;3(1):24-29. 10.14791/btrt.2015.3.1.24.
Validation of Housekeeping Genes for Gene Expression Analysis in Glioblastoma Using Quantitative Real-Time Polymerase Chain Reaction
- Affiliations
-
- 1Department of Biotechnology, Dayananda Sagar College of Engineering, Bangalore, India. nraja7@gmail.com
- KMID: 1882006
- DOI: http://doi.org/10.14791/btrt.2015.3.1.24
Abstract
- BACKGROUND
Quantitative real-time polymerase chain reaction (qPCR) is the most reliable tool for gene expression studies. Selection of housekeeping genes (HKGs) that are having most stable expression is critical to carry out accurate gene expression profiling. There is no 'universal' HKG having stable expression in all kinds of tissues under all experimental conditions.
METHODS
The present study aims to identify most appropriate HKGs for gene expression analysis in glioblastoma (GBM) samples. Based on literature survey, six most commonly used HKGs that are invariant in GBM were chosen. We performed qPCR using RNA from formalin fixed paraffin embedded GBM samples and normal brain samples to investigate the expression pattern of HPRT, GAPDH, TBP, B2M, B2M, RPL13A, and RN18S1 with different abundance. A simple Deltacycle threshold approach was employed to calculate the fold change.
RESULTS
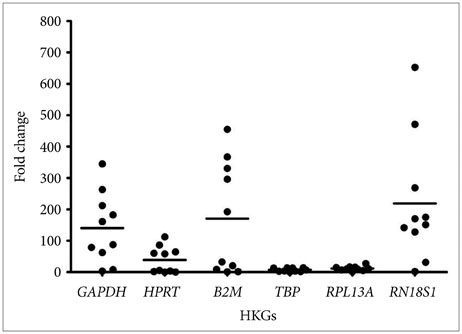
Our study shows that the expression of RPL13A and TBP were found to be most stable across all the samples and are thus suitable for gene expression analysis in human GBM. Except for TBP, none of the other conventionally used HKGs in GBM studies e.g., HPRT and GAPDH were found to be suitable as they showed variation in RNA expression.
CONCLUSION
Validation of HKGs is therefore immensely specific for a particular experimental setup and is crucial in assessing any new setup.
MeSH Terms
Figure
Reference
-
1. Ginzinger DG. Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp Hematol. 2002; 30:503–512.2. Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol. 2002; 29:23–39.
Article3. Kubista M, Andrade JM, Bengtsson M, et al. The real-time polymerase chain reaction. Mol Aspects Med. 2006; 27:95–125.
Article4. Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005; 6:279–284.
Article5. Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. Biotechniques. 2005; 39:75–85.
Article6. Dheda K, Huggett JF, Chang JS, et al. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal Biochem. 2005; 344:141–143.
Article7. Bonefeld BE, Elfving B, Wegener G. Reference genes for normalization: a study of rat brain tissue. Synapse. 2008; 62:302–309.
Article8. de Kok JB, Roelofs RW, Giesendorf BA, et al. Normalization of gene expression measurements in tumor tissues: comparison of 13 endogenous control genes. Lab Invest. 2005; 85:154–159.
Article9. Valente V, Teixeira SA, Neder L, et al. Selection of suitable housekeeping genes for expression analysis in glioblastoma using quantitative RT-PCR. BMC Mol Biol. 2009; 10:17.
Article10. Barber RD, Harmer DW, Coleman RA, Clark BJ. GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol Genomics. 2005; 21:389–395.
Article11. Cai J, Li T, Huang B, et al. The use of laser microdissection in the identification of suitable reference genes for normalization of quantitative real-time PCR in human FFPE epithelial ovarian tissue samples. PLoS One. 2014; 9:e95974.
Article12. Li R, Shen Y. An old method facing a new challenge: re-visiting housekeeping proteins as internal reference control for neuroscience research. Life Sci. 2013; 92:747–751.
Article13. Klatte M, Bauer P. Accurate Real-time Reverse Transcription Quantitative PCR. Methods Mol Biol. 2009; 479:61–77.
Article14. Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006; 1:1559–1582.
Article15. Radonić A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004; 313:856–862.
Article16. Tricarico C, Pinzani P, Bianchi S, et al. Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal Biochem. 2002; 309:293–300.
Article17. Chervoneva I, Li Y, Schulz S, et al. Selection of optimal reference genes for normalization in quantitative RT-PCR. BMC Bioinformatics. 2010; 11:253.
Article18. Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002; 3:RESEARCH0034.19. Colman H, Aldape K. Molecular predictors in glioblastoma: toward personalized therapy. Arch Neurol. 2008; 65:877–883.20. Zhou YH, Hess KR, Liu L, Linskey ME, Yung WK. Modeling prognosis for patients with malignant astrocytic gliomas: quantifying the expression of multiple genetic markers and clinical variables. Neuro Oncol. 2005; 7:485–494.
Article21. Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007; 114:97–109.
Article22. Louis DN. Molecular pathology of malignant gliomas. Annu Rev Pathol. 2006; 1:97–117.
Article23. Omuro AM, Faivre S, Raymond E. Lessons learned in the development of targeted therapy for malignant gliomas. Mol Cancer Ther. 2007; 6:1909–1919.
Article24. Chari R, Lonergan KM, Pikor LA, et al. A sequence-based approach to identify reference genes for gene expression analysis. BMC Med Genomics. 2010; 3:32.
Article25. Schmid H, Cohen CD, Henger A, Irrgang S, Schlöndorff D, Kretzler M. Validation of endogenous controls for gene expression analysis in microdissected human renal biopsies. Kidney Int. 2003; 64:356–360.
Article26. Curtis KM, Gomez LA, Rios C, et al. EF1alpha and RPL13a represent normalization genes suitable for RT-qPCR analysis of bone marrow derived mesenchymal stem cells. BMC Mol Biol. 2010; 11:61.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Reference gene selection for gene expression study in tissues of long-tailed chickens
- Real-time Polymerase Chain Reaction (PCR) in Microbiology
- Gene Amplification and Overexpression in BPH by Means of One-step Real Time Quantitative PCR
- Identification of Up-Regulated Genes in Malignant Glioma with Subtraction Hybridization: Preliminary Screening Studies
- Gene Expression in Cultured Keloid Fibroblasts