Ann Lab Med.
2014 Mar;34(2):134-138. 10.3343/alm.2014.34.2.134.
Identification of a De Novo Heterozygous Missense FLNB Mutation in Lethal Atelosteogenesis Type I by Exome Sequencing
- Affiliations
-
- 1Department of Pediatrics, Inje University College of Medicine, Busan Paik Hospital, Busan, Korea.
- 2Department of Laboratory Medicine and Genetics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea. changski@skku.edu
- 3Department of Obstetrics and Gynecology, Catholic University of Daegu, Daegu Catholic University Medical Center, Daegu, Korea.
- 4Department of Obstetrics and Gynecology, Inje University College of Medicine, Busan Paik Hospital, Busan, Korea.
- KMID: 1791906
- DOI: http://doi.org/10.3343/alm.2014.34.2.134
Abstract
- BACKGROUND
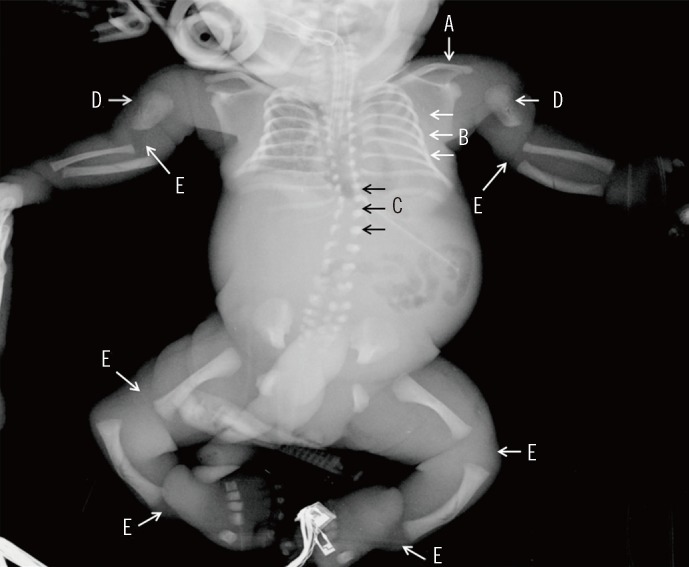
Atelosteogenesis type I (AO-I) is a rare lethal skeletal dysplastic disorder characterized by severe short-limbed dwarfism and dislocated hips, knees, and elbows. AO-I is caused by mutations in the filamin B (FLNB) gene; however, several other genes can cause AO-like lethal skeletal dysplasias.
METHODS
In order to screen all possible genes associated with AO-like lethal skeletal dysplasias simultaneously, we performed whole-exome sequencing in a female newborn having clinical features of AO-I.
RESULTS
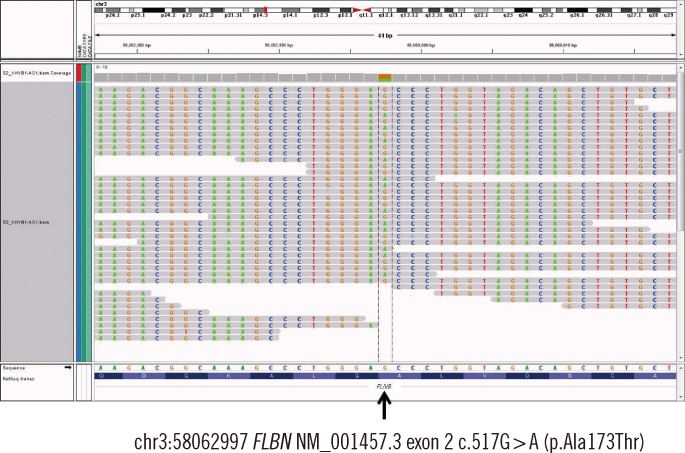
Exome sequencing identified a novel missense variant (c.517G>A; p.Ala173Thr) in exon 2 of the FLNB gene in the patient. Sanger sequencing validated this variant, and genetic analysis of the patient's parents suggested a de novo occurrence of the variant.
CONCLUSIONS
This study shows that exome sequencing can be a useful tool for the identification of causative mutations in lethal skeletal dysplasia patients.
Keyword
MeSH Terms
Figure
Reference
-
1. Lemyre E, Azouz EM, Teebi AS, Glanc P, Chen MF. Bone dysplasia series. Achondroplasia, hypochondroplasia and thanatophoric dysplasia: review and update. Can Assoc Radiol J. 1999; 50:185–197. PMID: 10405653.2. Maroteaux P, Spranger J, Stanescu V, Le Marec B, Pfeiffer RA, Beighton P, et al. Atelosteogenesis. Am J Med Genet. 1982; 13:15–25. PMID: 7137218.
Article3. Brodie SG, Lachman RS, Crandall BF, Fox MA, Rimoin DL, Cohn DH, et al. Radiographic and morphologic findings in a previously undescribed type of mesomelic dysplasia resembling atelosteogenesis type II. Am J Med Genet. 1998; 80:247–251. PMID: 9843047.
Article4. Verloes A, Lesenfants S, Barr M, Grange DK, Journel H, Lombet J, et al. Fronto-otopalatodigital osteodysplasia: clinical evidence for a single entity encompassing Melnick-Needles syndrome, otopalatodigital syndrome types 1 and 2, and frontometaphyseal dysplasia. Am J Med Genet. 2000; 90:407–422. PMID: 10706363.
Article5. Farrington-Rock C, Firestein MH, Bicknell LS, Superti-Furga A, Bacino CA, Cormier-Daire V, et al. Mutations in two regions of FLNB result in atelosteogenesis I and III. Hum Mutat. 2006; 27:705–710. PMID: 16752402.6. Mäkitie O, Savarirayan R, Bonafé L, Robertson S, Susic M, Superti-Furga A, et al. Autosomal recessive multiple epiphyseal dysplasia with homozygosity for C653S in the DTDST gene: double-layer patella as a reliable sign. Am J Med Genet A. 2003; 122A:187–192. PMID: 12966518.7. Robertson S. FLNB-Related Disorders. In : Pagon RA, Adam MP, Bird TD, Dolan CR, Fong CT, Stephens K, editors. GeneReviews. Seattle (WA): University of Washington, Seattle;1993-2013.8. Stern HJ, Graham JM Jr, Lachman RS, Horton W, Bernini PM, Spiegel PK, et al. Atelosteogenesis type III: a distinct skeletal dysplasia with features overlapping atelosteogenesis and oto-palato-digital syndrome type II. Am J Med Genet. 1990; 36:183–195. PMID: 2368807.
Article9. GeneTests. Accessed on Apr 30, 2013. http://www.genetests.org.10. Bettencourt C, López-Sendón J, García-Caldentey J, Rizzu P, Bakker I, Shomroni O, et al. Exome sequencing is a useful diagnostic tool for complicated forms of hereditary spastic paraplegia. Clin Genet. 2013; [Epub ahead of print].
Article11. Choi BO, Koo SK, Park MH, Rhee H, Yang SJ, Choi KG, et al. Exome sequencing is an efficient tool for genetic screening of Charcot-Marie-Tooth disease. Hum Mutat. 2012; 33:1610–1615. PMID: 22730194.
Article12. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010; 20:1297–1303. PMID: 20644199.
Article13. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variant discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011; 43:491–498. PMID: 21478889.14. Tekin M, Chioza BA, Matsumoto Y, Diaz-Horta O, Cross HE, Duman D, et al. SLITRK6 mutations cause myopia and deafness in humans and mice. J Clin Invest. 2013; 123:2094–2102. PMID: 23543054.15. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010; 38:e164. PMID: 20601685.
Article16. Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001; 29:308–311. PMID: 11125122.
Article17. Feng Y, Walsh CS. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol. 2004; 6:1034–1038. PMID: 15516996.
Article18. Dalkilic I, Schienda J, Thompson TG, Kunkel LM. Loss of FilaminC (FLNc) results in severe defects in myogenesis and myotube structure. Mol Cell Biol. 2006; 26:6522–6534. PMID: 16914736.
Article19. Hart AW, Morgan JE, Schneider J, West K, McKie L, Bhattacharya S, et al. Cardiac malformations and midline skeletal defects in mice lacking filamin A. Hum Mol Genet. 2006; 15:2457–2467. PMID: 16825286.
Article20. Temple K, Hall CA, Chitty L, Baraitser M. A case of atelosteogenesis. J Med Genet. 1990; 27:194–197. PMID: 2325095.
Article21. Bicknell LS, Morgan T, Bonafé L, Wessels MW, Bialer MG, Willems PJ, et al. Mutations in FLNB cause boomerang dysplasia. J Med Genet. 2005; 42:e43. PMID: 15994868.22. Bejjani BA, Oberg KC, Wilkins I, Moise A, Langston C, Superti-Furga A, et al. Prenatal ultrasonographic description and postnatal pathological findings in atelosteogenesis type 1. Am J Med Genet. 1998; 79:392–395. PMID: 9779808.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- De novo HCN1 Mutation Identified by Next-Generation Sequencing in a Patient with Early Infantile Epileptic Encephalopathy: Case Report
- Identification of a Novel De Novo Variant in the PAX3 Gene in Waardenburg Syndrome by Diagnostic Exome Sequencing: The First Molecular Diagnosis in Korea
- Identifying the KAT6B Mutation via Diagnostic Exome Sequencing to Diagnose Say-Barber-Biesecker-Young-Simpson Syndrome in Three Generations of a Family
- A family with X-linked Cornelia de Lange syndrome due to a novel SMC1A missense mutation identified by multi-gene panel sequencing
- Genetic Analysis of CLCN7 in an Old Female Patient with Type II Autosomal Dominant Osteopetrosis