Molecular Analysis of Isoleucyl-tRNA Synthetase Mutations in Clinical Isolates of Methicillin-Resistant Staphylococcus aureus with Low-Level Mupirocin Resistance
- Affiliations
-
- 1Division of Infectious Diseases*, Korea University College of Medicine, Korea. macropha@korea.ac.kr
- 2Research Institute of Emerging Infectious Diseases, Korea University, Seoul, Korea.
- 3Graduate School of Life Science and Biotechnology, Korea University, Seoul, Korea.
- KMID: 1781900
- DOI: http://doi.org/10.3346/jkms.2006.21.5.827
Abstract
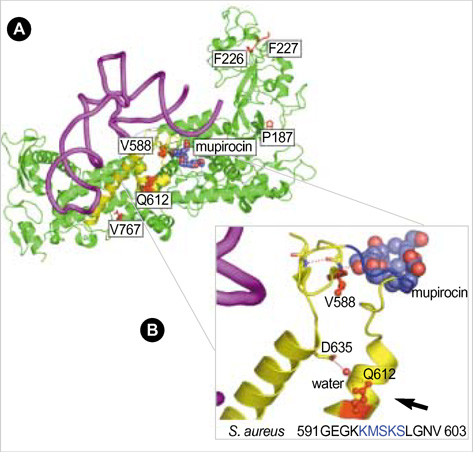
- Emergence and spread of low-level mupirocin resistance in staphylococci have been increasingly reported in recent years. The aim of this study was to characterize missense mutations within the chromosomal isoleucyl-tRNA synthetase gene (ileS) among clinical isolates of methicillin-resistant Staphylococcus aureus (MRSA) with low-level mupirocin resistance. A total of 20 isolates of MRSA with low-level mupirocin resistance (minimal inhibitory concentration, 16-64 microgram/mL) were collected from 79 patients in intensive care units for six months. The isolates were analyzed for isoleucyl-tRNA synthetase (IleS) mutations that might affect the binding of mupirocin to the three-dimensional structure of the S. aureus IleS enzyme. All isolates with low-level mupirocin resistance contained the known V588F mutation affecting the Rossman fold, and some of them additionally had previously unidentified mutations such as P187F, K226T, F227L, Q612H, or V767D. Interestingly, Q612H was a novel mutation that was involved in stabilizing the conformation of the catalytic loop containing the KMSKS motif. In conclusion, this study confirms that molecular heterogeneity in ileS gene is common among clinical MRSA isolates with low-level mupirocin resistance, and further study on clinical mutants is needed to understand the structural basis of low-level mupirocin resistance.
Keyword
MeSH Terms
Figure
Cited by 5 articles
-
The Prevalence, Genotype and Antimicrobial Susceptibility of High- and Low-Level Mupirocin Resistant Methicillin-Resistant Staphylococcus aureus
Se Young Park, Shin Moo Kim, Seok Don Park
Ann Dermatol. 2012;24(1):32-38. doi: 10.5021/ad.2012.24.1.32.The Prevalence, Genotype and Antimicrobial Susceptibility of High- and Low-Level Mupirocin Resistant Methicillin-Resistant Staphylococcus aureus
Se Young Park, Shin Moo Kim, Seok Don Park
Ann Dermatol. 2012;24(1):32-38. doi: 10.5021/ad.2012.24.1.32.In Vitro Antimicrobial Activities of Fusidic Acid and Retapamulin against Mupirocin- and Methicillin-Resistant Staphylococcus aureus
Sang Hyun Park, Jin Kyung Kim, Kun Park
Ann Dermatol. 2015;27(5):551-556. doi: 10.5021/ad.2015.27.5.551.Molecular Epidemiologic Study of a Methicillin-resistant Staphylococcus aureus Outbreak at a Newborn Nursery and Neonatal Intensive Care Unit
Hyun Mi Kang, Ki Cheol Park, Kyung-Yil Lee, Joonhong Park, Sun Hee Park, Dong-Gun Lee, Jong-Hyun Kim
Pediatr Infect Vaccine. 2019;26(3):148-160. doi: 10.14776/piv.2019.26.e23.Prevalence and Clinical Characteristics of Mupirocin-Resistant
Staphylococcus aureus
A-jin Lee, Hun-Suk Suh, Chang-Ho Jeon, Sang-Gyung Kim
Korean J Clin Microbiol. 2011;14(1):18-23. doi: 10.5145/KJCM.2011.14.1.18.
Reference
-
1. Muder RR, Brennen C, Wagener MM, Vickers RM, Rihs JD, Hancock GA, Yee YC, Miller JM, Yu VL. Methicillin-resistant staphylococcal colonization and infection in a long-term care facility. Ann Intern Med. 1991. 114:107–112.
Article2. Kluytmans J, van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev. 1997. 10:505–520.3. Rahman M, Noble WC, Cookson B, Baird D, Coia J. Mupirocin-resistant Staphylococcus aureus. Lancet. 1987. 2:387–388.4. Schmitz FJ, Lindenlauf E, Hofmann B, Fluit AC, Verhoef J, Heinz HP, Jones ME. The prevalence of low- and high-level mupirocin resistance in staphylococci from 19 European hospitals. J Antimicrob Chemother. 1998. 42:489–495.
Article5. Watanabe H, Masaki H, Asoh N, Watanabe K, Oishi K, Furumoto A, Kobayashi S, Sato A, Nagatake T. Emergence and spread of low-level mupirocin resistance in methicillin-resistant Staphylococcus aureus isolated from a community hospital in Japan. J Hosp Infect. 2001. 47:294–300.6. Gales AC, Andrade SS, Sader HS, Jones RN. Activity of mupirocin and 14 additional antibiotics against staphylococci isolated from Latin American hospitals: report from the SENTRY antimicrobial surveillance program. J Chemother. 2004. 16:323–328.
Article7. Yanagisawa T, Lee JT, Wu HC, Kawakami M. Relationship of protein structure of isoleucyl-tRNA synthetase with pseudomonic acid resistance of Escherichia coli. A proposed mode of action of pseudomonic acid as an inhibitor of isoleucyl-tRNA synthetase. J Biol Chem. 1994. 269:24304–24309.
Article8. Gilbart J, Perry CR, Slocombe B. High-level mupirocin resistance in Staphylococcus aureus: evidence for two distinct isoleucyl-tRNA synthetases. Antimicrob Agents Chemother. 1993. 37:32–38.9. Ramsey MA, Bradley SF, Kauffman CA, Morton TM. Identification of chromosomal location of mupA gene, encoding low-level mupirocin resistance in staphylococcal isolates. Antimicrob Agents Chemother. 1996. 40:2820–2823.
Article10. Yun HJ, Lee SW, Yoon GM, Kim SY, Choi S, Lee YS, Choi EC, Kim S. Prevalence and mechanisms of low- and high-level mupirocin resistance in staphylococci isolated from a Korean hospital. J Antimicrob Chemother. 2003. 51:619–623.
Article11. Walker ES, Vasquez JE, Dula R, Bullock H, Sarubbi FA. Mupirocin-resistant, methicillin-resistant Staphylococcus aureus: does mupirocin remain effective? Infect Control Hosp Epidemiol. 2003. 24:342–346.12. Antonio M, McFerran N, Pallen MJ. Mutations affecting the Rossman fold of isoleucyl-tRNA synthetase are correlated with low-level mupirocin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2002. 46:438–442.13. Fujimura S, Tokue Y, Watanabe A. Isoleucyl-tRNA synthetase mutations in Staphylococcus aureus clinical isolates and in vitro selection of low-level mupirocin-resistant strains. Antimicrob Agents Chemother. 2003. 47:3373–3374.14. National Committee for Clinical Laboratory Standards. Performance Standards for Antimicrobial Disk Susceptibility Tests. Approved Standard M2-M5. 1993. Villanova. Pa: National Committee for Laboratory Standards.15. National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically-fifty edition. Approved Standard M7-A5. 2000. Wayne, Pa: National Committee for Laboratory Standards.16. Silvian LF, Wang J, Steitz TA. Insights into editing from an ile-tRNA synthetase structure with tRNAile and mupirocin. Science. 1999. 285:1074–1077.17. Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000. 44:1549–1555.18. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995. 33:2233–2239.
Article19. Hurdle JG, O'Neill AJ, Ingham E, Fishwick C, Chopra I. Analysis of mupirocin resistance and fitness in Staphylococcus aureus by molecular genetic and structural modeling techniques. Antimicrob Agents Chemother. 2004. 48:4366–4376.20. Fujimura S, Watanabe A. Survey of high- and low-level mupirocin-resistant strains of methicillin-resistant Staphylococcus aureus in 15 Japanese hospitals. Chemotherapy. 2003. 49:36–38.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- In Vitro Antimicrobial Activities of Fusidic Acid and Retapamulin against Mupirocin- and Methicillin-Resistant Staphylococcus aureus
- Prevalence and Clinical Characteristics of Mupirocin-Resistant Staphylococcus aureus
- The Prevalence, Genotype and Antimicrobial Susceptibility of High- and Low-Level Mupirocin Resistant Methicillin-Resistant Staphylococcus aureus
- Isolation and Antimicrobial Susceptibility of Mupirocin-Resistant and Methicillin-Resistant Staphylococcus aureus from Clinical Samples
- Methicillin-resistant Staphylococcus aureus (MRSA) Infection in Neonates