Korean J Lab Med.
2011 Oct;31(4):282-284. 10.3343/kjlm.2011.31.4.282.
Comparison of an Automated Repetitive Sequence-based PCR Microbial Typing System with IS6110-Restriction Fragment Length Polymorphism for Epidemiologic Investigation of Clinical Mycobacterium tuberculosis Isolates in Korea
- Affiliations
-
- 1Department of Laboratory Medicine, School of Medicine, Pusan National University, Yangsan, Korea. cchl@pusan.ac.kr
- 2Department of Laboratory Medicine, Sanggye Paik Hospital, Inje University, Seoul, Korea.
- 3Department of Laboratory Medicine, University of Ulsan College of Medicine Ulsan University Hospital, Ulsan, Korea.
- 4Department of Laboratory Medicine, Dong-A University College of Medicine, Busan, Korea. jmkim@dau.ac.kr
- KMID: 1781693
- DOI: http://doi.org/10.3343/kjlm.2011.31.4.282
Abstract
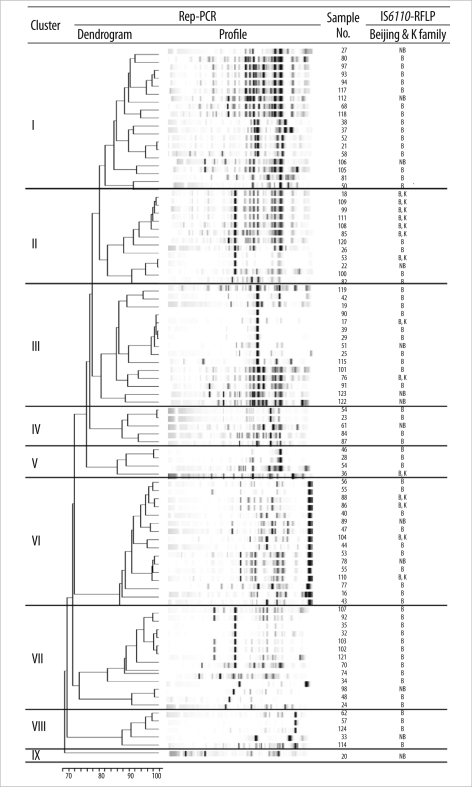
- Tuberculosis remains a severe public health problem worldwide. Presently, genotyping is used for conducting epidemiologic and clinical studies on tuberculosis cases. We evaluated the efficacy of the repetitive sequence-based PCR (rep-PCR)-based DiversiLab(TM) system (bioMerieux, France) over the IS6110-restriction fragment length polymorphism analysis for detecting Mycobacterium tuberculosis. In all, 89 clinical M. tuberculosis isolates collected nationwide from Korea were used. The DiversiLab system allocated the 89 isolates to 8 groups with 1 unique isolate when a similarity level of 95% was applied. Seventy-six isolates of the Beijing family and 13 isolates of non-Beijing family strains were irregularly distributed regardless of rep-PCR groups. The DiversiLab system generated a rapid, sensitive, and standardized result. It can be used to conduct molecular epidemiologic studies to identify clinical M. tuberculosis isolates in Korea.
Keyword
MeSH Terms
-
Automation
*Bacterial Typing Techniques
*Epidemiologic Methods
Genotype
Humans
Mycobacterium tuberculosis/*classification/genetics/isolation & purification
*Polymerase Chain Reaction
Polymorphism, Restriction Fragment Length
Reagent Kits, Diagnostic
Repetitive Sequences, Nucleic Acid
Republic of Korea/epidemiology
Tuberculosis/diagnosis/*epidemiology/microbiology
Figure
Reference
-
1. Global tuberculosis control: key findings from the December 2009 WHO report. Wkly Epidemiol Rec. 2010; 85:69–80. PMID: 20210259.2. Lönnroth K, Raviglione M. Global epidemiology of tuberculosis: prospects for control. Semin Respir Crit Care Med. 2008; 29:481–491. PMID: 18810682.
Article3. Espinal MA, Laszlo A, Simonsen L, Boulahbal F, Kim SJ, Reniero A, et al. Global trends in resistance to antituberculosis drugs. World Health Organization-International Union against Tuberculosis and Lung Disease Working Group on Anti-Tuberculosis Drug Resistance Surveillance. N Engl J Med. 2001; 344:1294–1303. PMID: 11320389.4. Gopaul KK, Brown TJ, Gibson AL, Yates MD, Drobniewski FA. Progression toward an improved DNA amplification-based typing technique in the study of Mycobacterium tuberculosis epidemiology. J Clin Microbiol. 2006; 44:2492–2498. PMID: 16825370.5. Dombek PE, Johnson LK, Zimmerley ST, Sadowsky MJ. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl Environ Microbiol. 2000; 66:2572–2577. PMID: 10831440.6. Healy M, Huong J, Bittner T, Lising M, Frye S, Raza S, et al. Microbial DNA typing by automated repetitive-sequence-based PCR. J Clin Microbiol. 2005; 43:199–207. PMID: 15634972.
Article7. Carretto E, Barbarini D, Farina C, Grosini A, Nicoletti P, Manso E. Use of the DiversiLab semiautomated repetitive-sequence-based polymerase chain reaction for epidemiologic analysis on Acinetobacter baumannii isolates in different Italian hospitals. Diagn Microbiol Infect Dis. 2008; 60:1–7. PMID: 17888611.8. van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993; 31:406–409. PMID: 8381814.9. Choi GE, Jang MH, Song EJ, Jeong SH, Kim JS, Lee WG, et al. IS6110-restriction fragment length polymorphism and spoligotyping analysis of Mycobacterium tuberculosis clinical isolates for investigating epidemiologic distribution in Korea. J Korean Med Sci. 2010; 25:1716–1721. PMID: 21165284.10. Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988; 26:2465–2466. PMID: 3069867.
Article11. Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991; 19:6823–6831. PMID: 1762913.12. Versalovic J, Kapur V, Mason EO Jr, Shah U, Koeuth T, Lupski JR, et al. Penicillin-resistant Streptococcus pneumoniae strains recovered in Houston: identification and molecular characterization of multiple clones. J Infect Dis. 1993; 167:850–856. PMID: 8450250.13. Versalovic J, Kapur V, Koeuth T, Mazurek GH, Whittam TS, Musser JM, et al. DNA fingerprinting of pathogenic bacteria by fluorophore-enhanced repetitive sequence-based polymerase chain reaction. Arch Pathol Lab Med. 1995; 119:23–29. PMID: 7802548.14. Cangelosi GA, Freeman RJ, Lewis KN, Livingston-Rosanoff D, Shah KS, Milan SJ, et al. Evaluation of a high-throughput repetitive-sequence-based PCR system for DNA fingerprinting of Mycobacterium tuberculosis and Mycobacterium avium complex strains. J Clin Microbiol. 2004; 42:2685–2693. PMID: 15184453.15. van Soolingen D, Qian L, de Haas PE, Douglas JT, Traore H, Portaels F, et al. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J Clin Microbiol. 1995; 33:3234–3238. PMID: 8586708.16. Shamputa IC, Lee J, Allix-Béguec C, Cho EJ, Lee JI, Rajan V, et al. Genetic diversity of Mycobacterium tuberculosis isolates from a tertiary care tuberculosis hospital in South Korea. J Clin Microbiol. 2010; 48:387–394. PMID: 20018816.17. Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, Willery E, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2006; 44:4498–4510. PMID: 17005759.18. Bouakaze C, Keyser C, de Martino SJ, Sougakoff W, Veziris N, Dabernat H, et al. Identification and genotyping of Mycobacterium tuberculosis complex species by use of a SNaPshot Minisequencing-based assay. J Clin Microbiol. 2010; 48:1758–1766. PMID: 20220173.19. Choi GE, Jang MH, Cho HJ, Lee SM, Yi J, Lee EY, et al. Application of single-nucleotide polymorphism and mycobacterial interspersed repetitive units-variable number of tandem repeats analyses to clinical Mycobacterium tuberculosis isolates from Korea. Korean J Lab Med. 2011; 31:37–43. PMID: 21239869.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Molecular fingerprinting of clinical isolates of Mycobacterium bovis and Mycobacterium tuberculosis from India by restriction fragment length polymorphism (RFLP)
- Molecular Strain Typing of Mycobacterium tuberculosis: a Review of Frequently Used Methods
- IS6110 - RFLP Analysis using Mycobacterium tuberculosis Isolates from Taegu - Kyungpook Region
- Optimal Combination of VNTR Typing for Discrimination of Isolated Mycobacterium tuberculosis in Korea
- The Present and Future of Molecular Epidemiology in Tuberculosis