Korean J Lab Med.
2007 Oct;27(5):351-354. 10.3343/kjlm.2007.27.5.351.
A Case of Bacteremia by Atopobium rimae in a Patient with Liver Cirrhosis
- Affiliations
-
- 1Department of Laboratory Medicine, University of Ulsan College of Medicine and Asan Medical Center, Seoul, Korea. sung@amc.seoul.kr
- 2Department of Internal Medicine, University of Ulsan College of Medicine and Asan Medical Center, Seoul, Korea.
- KMID: 1781534
- DOI: http://doi.org/10.3343/kjlm.2007.27.5.351
Abstract
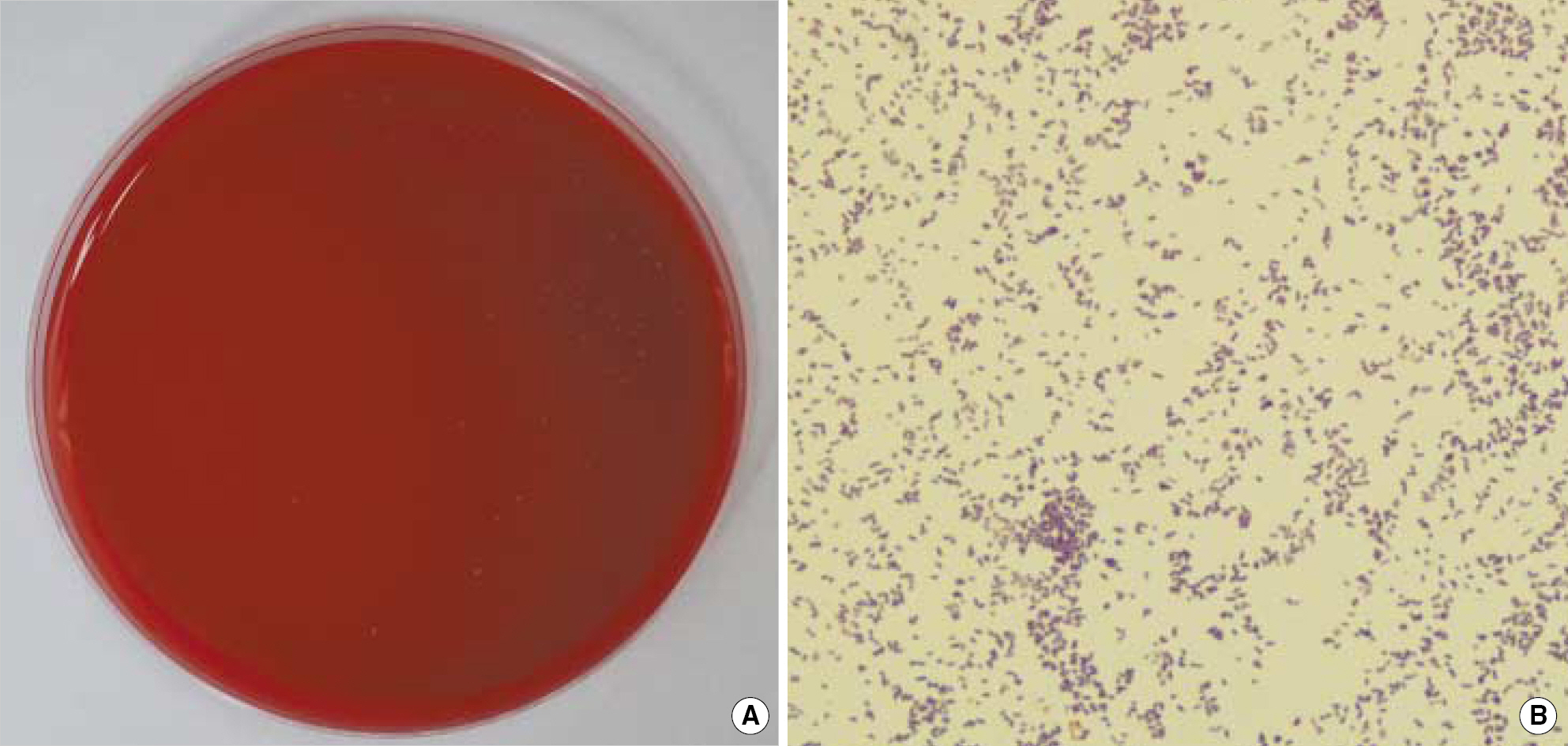
- Atopobium rimae, previously Lactobacillus rimae, is a strictly anaerobic, non-spore forming grampositive rod which was frequently isolated from odontogenic infection. We report a case of A. rimae bacteremia. A 47-yr-old man with liver cirrhosis was admitted to the hospital via emergency room due to fever and chill. His abdominal and pelvic computed tomography revealed a small abscess near the left adrenal gland. Three sets of blood cultures were taken and non-spore forming, grampositive rods were detected in all anaerobic vials. This isolate grew small nonhemolytic, gray-white translucent colonies on Brucella blood agar and was obligatory anaerobic on air-tolerance test. This organism was negative for catalase, indole, nitrate-reduction and beta-lactamase and failed to identify by Vitek ANI card (bioMerieux, France). 16S rRNA sequences of this showed 99.8% homology of the published sequence of A. rimae (GenBank accession number AF292371). Aspirates of periadrenal abscess grew Escherichia coli and Peptostreptococcus micros. He was treated with metronidazole and imipenem and follow-up cultures of blood were negative at days 4 and 10. To our knowledge, this is the first report of bacteremia of A. rimae.
Keyword
MeSH Terms
Figure
Reference
-
1.Kumar PS., Griffen AL., Barton JA., Paster BJ., Moeschberger ML., Leys EJ. New bacterial species associated with chronic periodontitis. J Dent Res. 2003. 82:338–44.
Article2.Konone E., Wade WG. Propionibacterium, Lactobacillus, Actinomyces, and other non-spore-forming anaerobic gram-positive rods. Murray PR, Baron EJ, editors. Manual of clinical microbiology. 9th ed.Washington, DC: ASM Press;2007. p. 872–88.3.Lim SH., Kim KW., Yoo SY., Kook JK., Chang YI. Identification of bacteria from the peri-implant sulcus of orthodontic mini-implants using 16S rDNA clone library. Korean J Orthod. 2006. 36:251–62. (임성훈, 김광원, 유소영, 국중기, 장영일. 16S rDNA 클론 library 제작 및핵산염기서열 결정을 통한 교정용 미니임플랜트 주위 열구의 세균 동정. 대한치과교정학회지 2006;36: 251-62.).4.Relman DA. Universal bacterial 16S rRNA amplification and sequencing. Persing DH, Smith TF, editors. Diagnostic molecular microbiology principles and applications. 1st ed.Washington DC: ASM Press;1993. p. 489–95.5.Citron DM., Hecht DW. Susceptibility test methods: anaerobic bacteria. Murray PR, Baron EJ, editors. Manual of clinical microbiology. 9th ed.Washington, DC: ASM Press;2007. p. 1214–22.6.Clinical and Laboratory Standards Institute. Methods for antimicrobial susceptibility testing of anaerobic bacteria. Approved Standard, M11-A7. Wayne, PA: Clinical and Laboratory Standards Institute;2007.7.Woo PC., Ng KH., Lau SK., Yip KT., Fung AM., Leung KW, et al. Usefulness of the MicroSeq 500 16S ribosomal DNA-based bacterial identification system for identification of clinically significant bacterial isolates with ambiguous biochemical profiles. J Clin Microbiol. 2003. 41:1996–2001.
Article8.Nolte FS., Caliendo AM. Molecular detection and identification of microorganisms. Murray PR, Baron EJ, editors. Manual of clinical microbiology. 9th ed.Washington, DC: ASM Press;2007. p. 218–44.9.Olsen I., Johnson JL., Moore LV., Moore WE. Lactobacillus uli sp. nov. and Lactobacillus rimae sp. nov. from the human gingival crevice and emended descriptions of Lactobacillus minutus and Streptococcus parvulus. Int J Syst Bacteriol. 1991. 41:261–6.10.Collins MD., Wallbanks S. Comparative sequence analyses of the 16S rRNA genes of Lactobacillus minutus, Lactobacillus rimae and Streptococcus parvulus: proposal for the creation of a new genus Atopobium. FEMS Microbiol Lett. 1992. 74:235–40.11.Kageyama A., Benno Y., Nakase T. Phylogenic and phenotypic evidence for the transfer of Eubacterium fossor to the genus Atopobium as Atopobium fossor comb. nov. Microbiol Immunol. 1999. 43:389–95.12.Rodriguez Jovita M., Collins MD., Sjoden B., Falsen E. Characterization of a novel Atopobium isolate from the human vagina: description of Atopobium vaginae sp. nov. Int J Syst Bacteriol. 1999. 49:1573–6.13.De Backer E., Verhelst R., Verstraelen H., Claeys G., Verschraegen G., Temmerman M, et al. Antibiotic susceptibility of Atopobium vaginae. BMC Infect Dis. 2006. 6:51.14.Ferris MJ., Masztal A., Aldridge KE., Fortenberry JD., Fidel PL Jr., Martin DH. Association of Atopobium vaginae, a recently described metronidazole resistant anaerobe, with bacterial vaginosis. BMC Infect Dis. 2004. 4:5.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A case of Bacteremia Caused by Vibrio parahaemolyticus in Liver Cirrhosis
- A Case of Streptococcus gallolyticus subsp. gallolyticus Bacteremia in Liver Cirrhosis
- Campylobacter jejuni Bacteremia in a Liver Cirrhosis Patient and Review of Literature: A Case Study
- A case of pyomyositis caused by Vibrio cholerae non-O1 in a patient with liver cirrhosis
- Two Cases of Bacteremia Caused by Vibrio Parahemolyticus in Liver Cirrhosis