Yonsei Med J.
2012 Jan;53(1):132-137. 10.3349/ymj.2012.53.1.132.
A Modified Extraction Method of Circulating Free DNA for Epidermal Growth Factor Receptor Mutation Analysis
- Affiliations
-
- 1Department of Oncology, No. 3 People's Hospital, School of Medicine, Shanghai Jiaotong University, Shanghai, China. dr_jiang@yeah.net
- 2Department of Oncology, No.113 Hospital of People's Liberation Army, Ningbo, China.
- 3Tumor Genetics Capability, Innovation Center China, Astrazeneca Global R&D, Shanghai, China.
- 4Department of Nuclear Medicine, Renji Hospital, School of Medicine, Shanghai Jiaotong University, Shanghai, China.
- KMID: 1779697
- DOI: http://doi.org/10.3349/ymj.2012.53.1.132
Abstract
- PURPOSE
Circulating free DNA (cfDNA) in plasma is promising to be a surrogate for tumor tissue DNA. However, not all epidermal growth factor receptor (EGFR) mutations in tumor tissue DNA has been detected in matched cfDNA, at least partly due to inefficient cfDNA extraction method. The purpose of this study was to establish an efficient plasma cfDNA extraction protocol.
MATERIALS AND METHODS
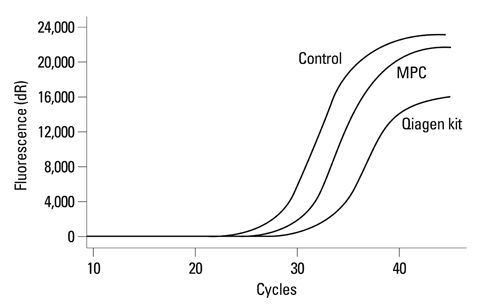
The yield of plasma cfDNA extracted by our modified phenol-chloroform (MPC) method from non-small-cell lung cancer (NSCLC) patients was compared with that by QIAamp MinElute Virus Spin kit (Qiagen kit) as control, using the Wilcoxon rank-sum test. TaqMan quantitative polymerase chain reaction (qPCR) assays were used to quantify the plasma cfDNA extracted. Both Mutant-enriched PCR (ME-PCR) coupled sequencing and DxS EGFR mutation test kit were used to evaluate the impact of extraction method on EGFR mutation analysis.
RESULTS
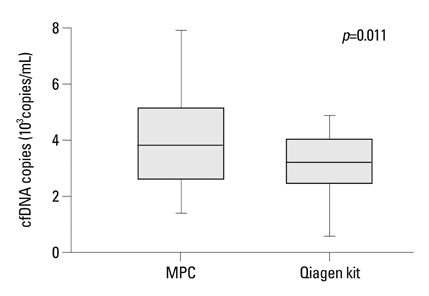
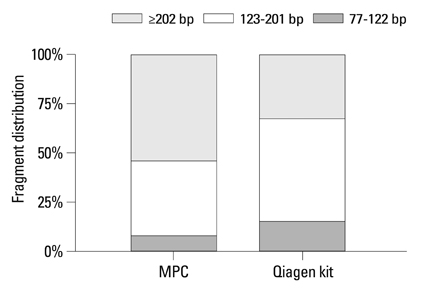
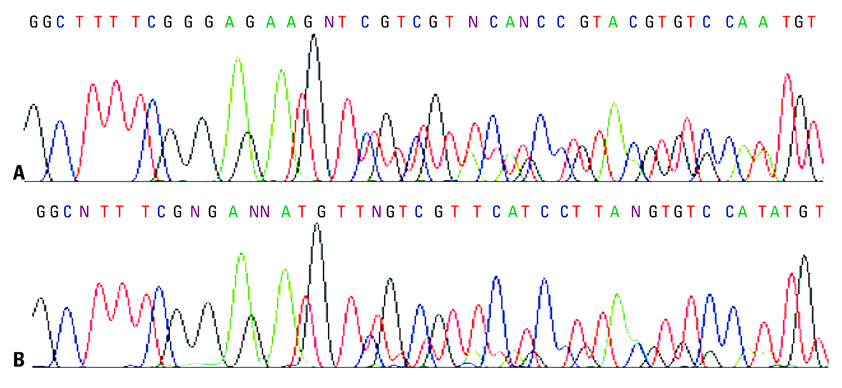
MPC method extracted more plasma cfDNA than Qiagen kit method (p=0.011). The proportion of longer fragment (> or =202 bp) in cfDNA extracted by MPC method was significantly higher than by Qiagen kit method (p=0.002). In the sequencing maps of ME-PCR products, a higher mutant peak was observed on plasma cfDNA extracted by MPC method than by Qiagen kit method. In DxS EGFR mutation test kit results, plasma cfDNA extracted by MPC method contained more tumor-origin DNA than by Qiagen kit method.
CONCLUSION
An improved plasma cfDNA extraction method of MPC is provided, which will be beneficial for EGFR mutation analysis for patients with NSCLC.
Keyword
MeSH Terms
Figure
Reference
-
1. Alberg AJ, Samet JM. Epidemiology of lung cancer. Chest. 2003. 123:21S–49S.
Article2. Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002. 346:92–98.
Article3. Jänne PA, Engelman JA, Johnson BE. Epidermal growth factor receptor mutations in non-small-cell lung cancer: implications for treatment and tumor biology. J Clin Oncol. 2005. 23:3227–3234.
Article4. Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009. 361:947–957.
Article5. Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007. 7:169–181.
Article6. Riely GJ, Politi KA, Miller VA, Pao W. Update on epidermal growth factor receptor mutations in non-small cell lung cancer. Clin Cancer Res. 2006. 12:7232–7241.
Article7. Bai H, Mao L, Wang HS, Zhao J, Yang L, An TT, et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J Clin Oncol. 2009. 27:2653–2659.
Article8. He C, Liu M, Zhou C, Zhang J, Ouyang M, Zhong N, et al. Detection of epidermal growth factor receptor mutations in plasma by mutant-enriched PCR assay for prediction of the response to gefitinib in patients with non-small-cell lung cancer. Int J Cancer. 2009. 125:2393–2399.
Article9. Kuang Y, Rogers A, Yeap BY, Wang L, Makrigiorgos M, Vetrand K, et al. Noninvasive detection of EGFR T790M in gefitinib or erlotinib resistant non-small cell lung cancer. Clin Cancer Res. 2009. 15:2630–2636.
Article10. Board RE, Williams VS, Knight L, Shaw J, Greystoke A, Ranson M, et al. Isolation and extraction of circulating tumor DNA from patients with small cell lung cancer. Ann N Y Acad Sci. 2008. 1137:98–107.
Article11. Kimura H, Kasahara K, Kawaishi M, Kunitoh H, Tamura T, Holloway B, et al. Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non-small-cell lung cancer. Clin Cancer Res. 2006. 12:3915–3921.
Article12. Kimura H, Suminoe M, Kasahara K, Sone T, Araya T, Tamori S, et al. Evaluation of epidermal growth factor receptor mutation status in serum DNA as a predictor of response to gefitinib (IRESSA). Br J Cancer. 2007. 97:778–784.
Article13. Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001. 61:1659–1665.14. Suzuki N, Kamataki A, Yamaki J, Homma Y. Characterization of circulating DNA in healthy human plasma. Clin Chim Acta. 2008. 387:55–58.
Article15. Lee TH, Montalvo L, Chrebtow V, Busch MP. Quantitation of genomic DNA in plasma and serum samples: higher concentrations of genomic DNA found in serum than in plasma. Transfusion. 2001. 41:276–282.
Article16. Taback B, O'Day SJ, Hoon DS. Quantification of circulating DNA in the plasma and serum of cancer patients. Ann N Y Acad Sci. 2004. 1022:17–24.
Article17. van der Drift MA, Hol BE, Klaassen CH, Prinsen CF, van Aarssen YA, Donders R, et al. Circulating DNA is a non-invasive prognostic factor for survival in non-small cell lung cancer. Lung Cancer. 2010. 68:283–287.
Article18. Yoon KA, Park S, Lee SH, Kim JH, Lee JS. Comparison of circulating plasma DNA levels between lung cancer patients and healthy controls. J Mol Diagn. 2009. 11:182–185.
Article19. Müller I, Beeger C, Alix-Panabières C, Rebillard X, Pantel K, Schwarzenbach H. Identification of loss of heterozygosity on circulating free DNA in peripheral blood of prostate cancer patients: potential and technical improvements. Clin Chem. 2008. 54:688–696.
Article20. Wang M, Block TM, Steel L, Brenner DE, Su YH. Preferential isolation of fragmented DNA enhances the detection of circulating mutated k-ras DNA. Clin Chem. 2004. 50:211–213.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Amplification of epidermal growth factor receptor gene in primary cervical cancer
- Amplification of epidermal growth factor receptor gene in primary cervical cancer
- Epidermal growth factor receptors increase in rabbit embryonal implantation
- Correlation of epidermal growth factor receptor expression with prognostic factors in patients with ovarian neoplasms
- Prognostic significance of epidermal growth factor receptor expression in human gastric carcinoma