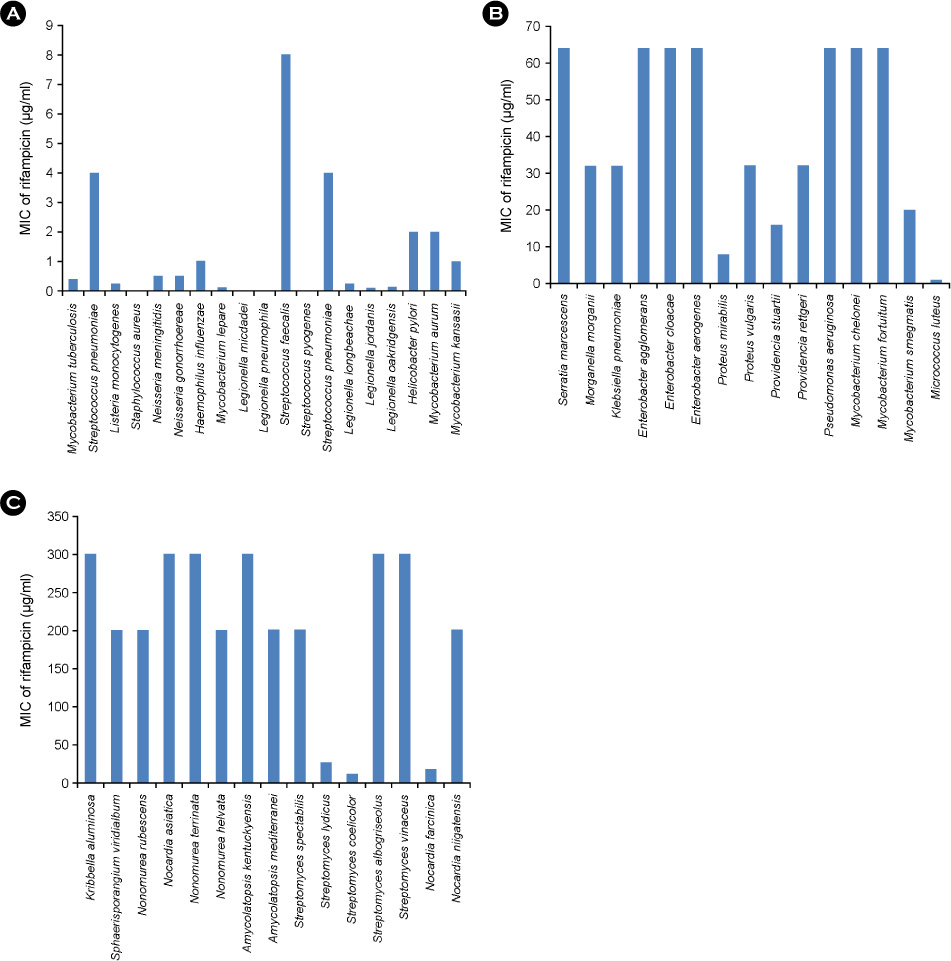
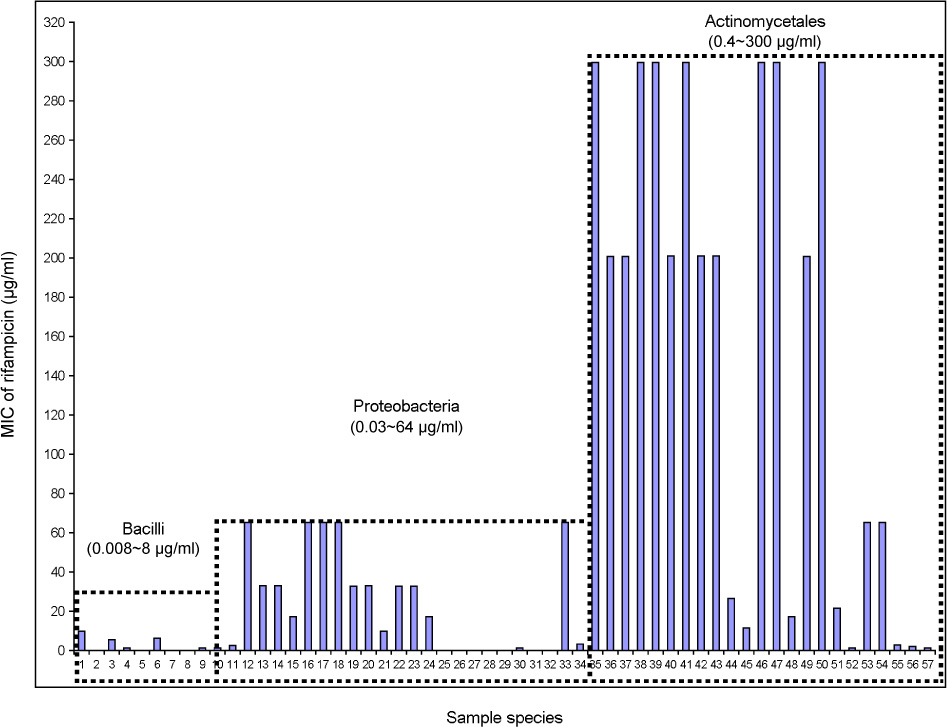
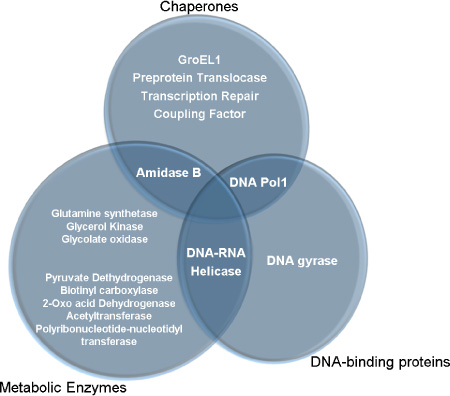
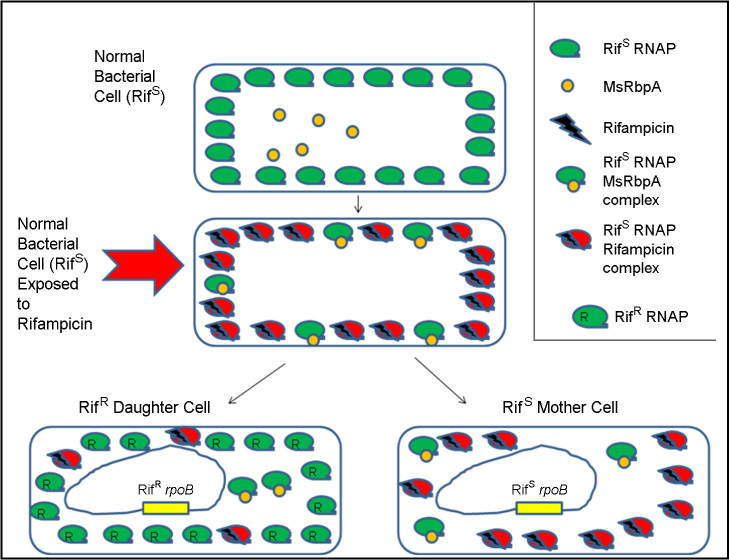
J Bacteriol Virol.
2012 Jun;42(2):87-100. 10.4167/jbv.2012.42.2.87.
Tracing the Variation in Physiological Response to Rifampicin Across the Microbial Spectrum
- Affiliations
-
- 1Molecular Biophysics Unit, Indian Institute of Science, Bangalore, India. dipankar@mbu.iisc.ernet.in
- KMID: 1717680
- DOI: http://doi.org/10.4167/jbv.2012.42.2.87
Abstract
- Rifampicin is an acknowledged inhibitor of bacterial RNA polymerase. We observed that there exists a substantial variation in the basal-level tolerance to rifampicin across microbial species. A number of mechanisms have come to light which depict the molecular aspects of the behavior of an individual bacterial cell towards rifampicin. This review assesses the current knowledge about rifampicin and conjectures about the probable aspects of rifampicin which remain unexplored.
Keyword
MeSH Terms
Figure
Reference
-
1. Darst SA. New inhibitors targeting bacterial RNA polymerase. Trends Biochem Sci. 2004. 29:159–160.
Article2. Quan S, Venter H, Dabbs ER. Ribosylative inactivation of rifampin by Mycobacterium smegmatis is a principal contributor to its low susceptibility to this antibiotic. Antimicrob Agents Chemother. 1997. 41:2456–2460.
Article3. Hu Y, Mangan JA, Dhillon J, Sole KM, Mitchison DA, Butcher PD, et al. Detection of mRNA transcripts and active transcription in persistent Mycobacterium tuberculosis induced by exposure to rifampin or pyrazinamide. J Bacteriol. 2000. 182:6358–6365.
Article4. Wegrzyn A, Szalewska-Palasz A, Blaszczak A, Liberek K, Wegrzyn G. Differential inhibition of transcription from δ70- and δ32-dependent promoters by rifampicin. FEBS Lett. 1998. 440:172–174.
Article5. Newell KV, Thomas DP, Brekasis D, Paget MS. The RNA polymerase-binding protein RbpA confers basal levels of rifampicin resistance on Streptomyces coelicolor. Mol Microbiol. 2006. 60:687–696.
Article6. Flåtten I, Morigen , Skarstad K. DnaA protein interacts with RNA polymerase and partially protects it from the effect of rifampicin. Mol Microbiol. 2009. 71:1018–1030.
Article7. Martinez JL. Antibiotics and antibiotic resistance genes in natural environments. Science. 2008. 321:365–367.
Article8. Oppolzer W, Prelog V, Sensi P. The composition of rifamycin B and related rifamycins. Experientia. 1964. 20:336–339.9. Hinkle DC, Mangel WF, Chamberlin MJ. Studies of the binding of Escherichia coli RNA polymerase to DNA. IV. The effect of rifampicin on binding and on RNA chain initiation. J Mol Biol. 1972. 70:209–220.10. Yarbrough LR, Wu FY, Wu CW. Molecular mechanism of the rifampicin -RNA polymerase interaction. Biochemistry. 1976. 15:2669–2676.
Article11. McClure WR, Cech CL. On the mechanism of rifampicin inhibition of RNA synthesis. J Biol Chem. 1978. 253:8949–8956.
Article12. Jin DJ, Gross CA. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988. 202:45–58.
Article13. Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, et al. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001. 104:901–912.
Article14. Korzheva N, Mustaev A, Kozlov M, Malhotra A, Nikiforov V, Goldfarb A, et al. A structural model of transcription elongation. Science. 2000. 289:619–625.
Article15. Thornsberry C, Hill BC, Swenson JM, McDougal LK. Rifampin: Spectrum of antibacterial activity. Rev Infect Dis. 1983. 5:S412–S417.
Article16. Williams KJ, Piddock LJ. Accumulation of rifampicin by Escherichia coli and Staphylococcus aureus. J Antimicrob Chemother. 1998. 42:597–603.
Article17. Hui J, Gordon N, Kajioka R. Permeability barrier to rifampin in mycobacteria. Antimicrob Agents Chemother. 1977. 11:773–779.
Article18. Siddiqi N, Das R, Pathak N, Banerjee S, Ahmed N, Katoch VM, et al. Mycobacterium tuberculosis isolate with a distinct genomic identity overexpresses a tap-like efflux pump. Infection. 2004. 32:109–111.
Article19. Hagman KE, Pan W, Spratt BG, Balthazar JT, Judd RC, Shafer WM. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology. 1995. 141:611–622.
Article20. Imai T, Watanabe K, Mikami Y, Yazawa K, Ando A, Nagata Y, et al. Identification and characterization of a new intermediate in the ribosylative inactivation pathway of rifampin by Mycobacterium smegmatis. Microb Drug Resist. 1999. 5:259–264.
Article21. Dabbs ER, Yazawa K, Tanaka Y, Mikami Y, Miyaji M, Andersen SJ, et al. Rifampicin inactivation by Bacillus species. J Antibiot. 1995. 48:815–819.22. Tanaka Y, Yazawa K, Dabbs ER, Nishikawa K, Komaki H, Mikami Y, et al. Different rifampicin inactivation mechanisms in Nocardia and related taxa. Microbiol Immunol. 1996. 40:1–4.
Article23. Ishikawa J, Yamashita A, Mikami Y, Hoshino Y, Kurita H, Hotta K, et al. The complete genome sequence of Nocardia farcinica IFM 10152. Proc Natl Acad Sci U S A. 2004. 101:14925–14930.24. Talà A, Wang G, Zemanova M, Okamoto S, Ochi K, Alifano P. Activation of dormant bacterial genes by Nonomuraea sp. strain ATCC 39727 mutant-type RNA polymerase. J Bacteriol. 2009. 191:805–814.
Article25. Kühner S, van Noort V, Betts MJ, Leo-Macias A, Batisse C, Rode M, et al. Proteome organization in a genome-reduced bacterium. Science. 2009. 326:1235–1240.
Article26. Mukherjee R, Chatterji D. Stationary phase induced alterations in mycobacterial RNA polymerase assembly: A cue to its phenotypic resistnce towards rifampicin. Biochem Biophys Res Commun. 2008. 369:899–904.
Article27. Dey A, Verma AK, Chatterji D. Role of an RNA polymerase interacting protein, MsRbpA, from Mycobacterium smegmatis in phenotypic tolerance to rifampicin. Microbiology. 2010. 156:873–883.
Article28. Dey A, Verma AK, Chatterji D. Molecular insights into the mechanism of phenotypic tolerance to rifampicin conferred on mycobacterial RNA polymerase by MsRbpA. Microbiology. 2011. 157:2056–2071.
Article29. Liu A, Tran L, Becket E, Lee K, Chinn L, Park E, et al. Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: Generating an antibiotic bar code. Antimicrob Agents Chemother. 2010. 54:1393–1403.
Article30. Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006. 2:2006.0008.31. Ren H, Liu J. AsnB is involved in natural resistance of Mycobacterium smegmatis to multiple drugs. Antimicrob Agents Chemother. 2006. 50:250–255.
Article32. Ojha A, Anand M, Bhatt A, Kremer L, Jacobs WR Jr, Hatfull GF. GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobateria. Cell. 2005. 123:861–873.
Article33. Kovacs L, Csanadi A, Megyeri K, Kaberdin VR, Miczak A. Mycobacterial RNase E-associated proteins. Microbiol Immunol. 2005. 49:1003–1007.
Article34. Macinga DR, Rather PN. aarD, a Providencia stuartii homologue of cydD: role in 2'-N-acetyltransferase expression, cell morphology and growth in the presence of an extracellular factor. Mol Microbiol. 1996. 19:511–520.
Article35. Handschin JC, Wehrli W. On the kinetics of the rifampicin-RNA-polymerase complex. Differences between crude and purified enzyme fractions. Eur J Biochem. 1976. 66:309–317.
Article36. Kumar KP, Reddy PS, Chatterji D. Proximity relationship between the active site of Escherichia coli RNA polymerase and rifampicin binding domain: A resonance energy-transfer study. Biochemistry. 1992. 31:7519–7526.
Article37. Harshey RM, Ramakrishnan T. Purification and properties of DNA-dependent RNA polymerase from Mycobacterium tuberculosis H37RV. Biochim Biophys Acta. 1976. 432:49–59.
Article38. Fábry M, Sümegi J, Venetianer P. Purification and properties of the RNA polymerase of an extremely thermophilic bacterium: Thermus aquaticus T2. Biochim Biophys Acta. 1976. 435:228–235.
Article39. Zenkin N, Kulbachinskiy A, Bass I, Nikiforov V. Different rifampin sensitivities of Escherichia coli and Mycobacterium tuberculosis RNA polymerases are not explained by the difference in the β-subunit rifampin regions I and II. Antimicrob Agents Chemother. 2005. 49:1587–1590.
Article40. Ishikawa J, Yamashita A, Mikami Y, Hoshino Y, Kurita H, Hotta K, et al. The complete genome sequence of Nocardia farcinica IFM 10152. Proc Natl Acad Sci U S A. 2004. 101:14925–14930.41. Vigliotta G, Tredici SM, Damiano F, Montinaro MR, Pulimeno R, di Summa R, et al. Natural merodiploidy involving duplicated rpoB alleles affects secondary metabolism in producer actinomycete. Mol Microbiol. 2005. 55:396–412.
Article42. Ishikawa J, Chiba K, Kurita H, Satoh H. Contribution of rpoB2 RNA polymerase β-subunit gene to rifampin resistance in Nocardia species. Antimicrob Agents Chemother. 2006. 50:1342–1346.
Article43. Zhou YN, Jin DJ. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like "stringent" RNA polymerases in Escherichia coli. Proc Natl Acad Sci U S A. 1998. 95:2908–2913.
Article44. Bandow JE, Brötz H, Hecker M. Bacillus subtilis tolerance of moderate concentrations of rifampin involves the δB-dependent general and multiple stress response. J Bacteriol. 2002. 184:459–467.
Article45. Aldridge BB, Fernandez-Suarez M, Heller D, Ambravaneswaran V, Irimia D, Toner M, et al. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science. 2012. 335:100–104.
Article46. Davies J, Spiegelman GB, Yim G. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol. 2006. 9:445–453.
Article47. Goh EB, Yim G, Tsui W, McClure J, Surette MG, Davies J. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc Natl Acad Sci U S A. 2002. 99:17025–17030.
Article48. Piddock LJ, Williams KJ, Ricci V. Accumulation of rifampicin by Mycobacterium aurum, Mycobacterium smegmatis and Mycobacterium tuberculosis. J Antimicrob Chemother. 2000. 45:159–165.
Article49. Dantas G, Sommer MO, Oluwasegun RD, Church GM. Bacteria subsisting on antibiotics. Science. 2008. 320:100–103.
Article50. Davies J. Darwin and microbiomes. EMBO Rep. 2009. 10:805.
Article51. Andersen SJ, Quan S, Gowan B, Dabbs ER. Monooxygenase-like sequence of a Rhodococcus equi gene conferring increased resistance torifampin by inactivating this antibiotic. Antimicrob Agents Chemother. 1997. 41:218–221.
Article52. Yazawa K, Mikami Y, Maeda A, Akao M, Morisaki N, Iwasaki S. Inactivation of rifampin by Nocardia brasiliensis. Antimicrob Agents Chemother. 1993. 37:1313–1317.53. Yazawa K, Mikami Y, Maeda A, Morisaki N, Iwasaki S. Phosphorylative inactivation of rifampicin by Nocardia otitidiscaviarum. J Antimicrob Chemother. 1994. 33:1127–1135.
Article54. Li XZ, Zhang L, Nikaido H. Efflux pump-mediated intrinsic drug resistance in Mycobacterium smegmatis. Antimicrob Agents Chemother. 2004. 48:2415–2423.
Article55. Eng RH, Padberg FT, Smith SM, Tan EN, Cherubin CE. Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob Agents Chemother. 1991. 35:1824–1828.
Article56. Xie Z, Siddiqi N, Rubin EJ. Differential antibiotic susceptibilities of starved Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother. 2005. 49:4778–4780.
Article57. Perkins AE, Nicholson WL. Uncovering new metabolic capabilities of Bacillus subtilis using phenotype profiling of rifampin-resistant rpoB mutants. J Bacteriol. 2008. 190:807–814.
Article58. Heep M, Odenbreit S, Beck D, Decker J, Prohaska E, Rieger U, et al. Mutations at four distinct regions of the rpoB gene can reduce the susceptibility of Helicobacter pylori to rifamycins. Antimicrob Agents Chemother. 2000. 44:1713–1715.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Comparative Analysis of Heart Rate Variability of Electrocardiogram and Pulse-wave Using Time Series
- Rifampicin Alleviates Atopic Dermatitis-Like Response in vivo and in vitro
- A Case of Rifampicin-induced Thrombocytopenic Purpura
- Reversible Acute Renal Failure Associated with Rifampicin: Report of a Case
- The Clinical Effect of Rifampicin for Retreatment Cases for Pulmonary Tuberculosis