J Vet Sci.
2013 Jun;14(2):115-124. 10.4142/jvs.2013.14.2.115.
Genetic diversity of porcine reproductive and respiratory syndrome virus in Korea
- Affiliations
-
- 1Animal and Plant Quarantine Agency, Ministry of Agriculture, Food and Rural Affairs, Anyang 430-757, Korea. shinyk2009@korea.kr
- 2College of Veterinary Medicine, Chonbuk National University, Jeonju 561-756, Korea.
- KMID: 1705512
- DOI: http://doi.org/10.4142/jvs.2013.14.2.115
Abstract
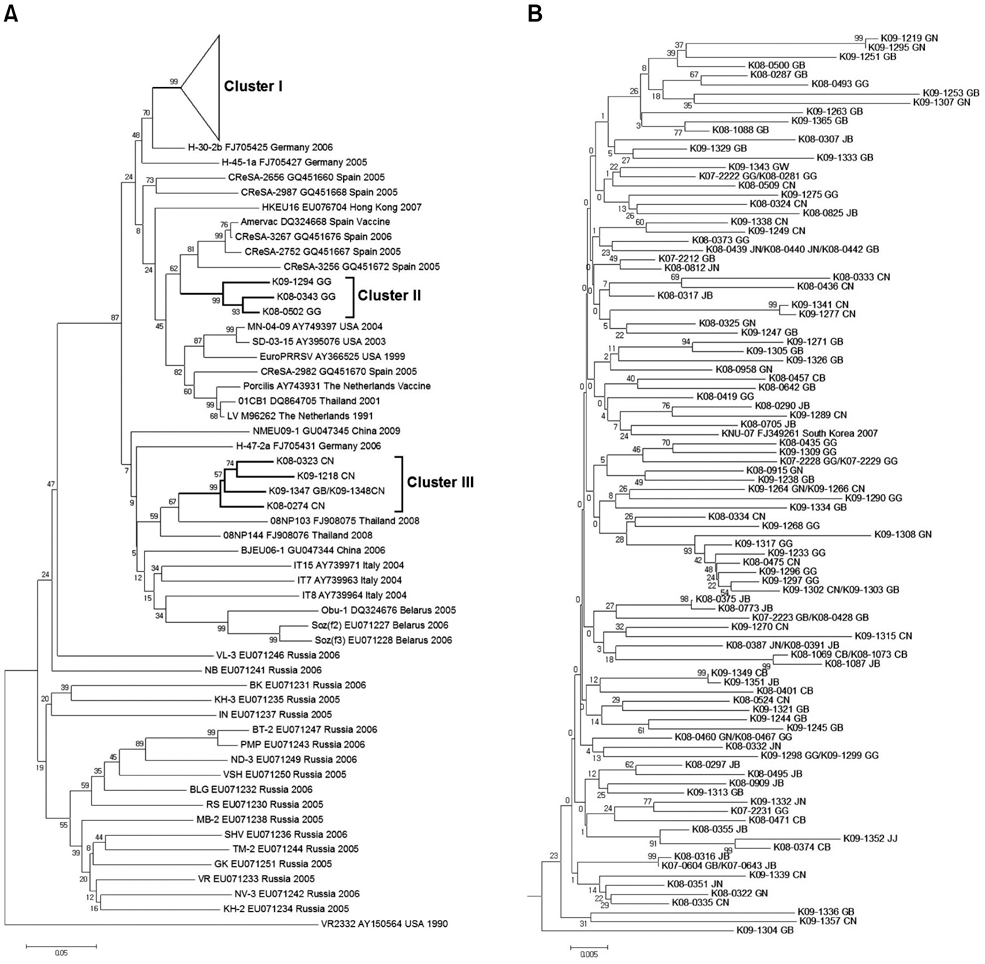
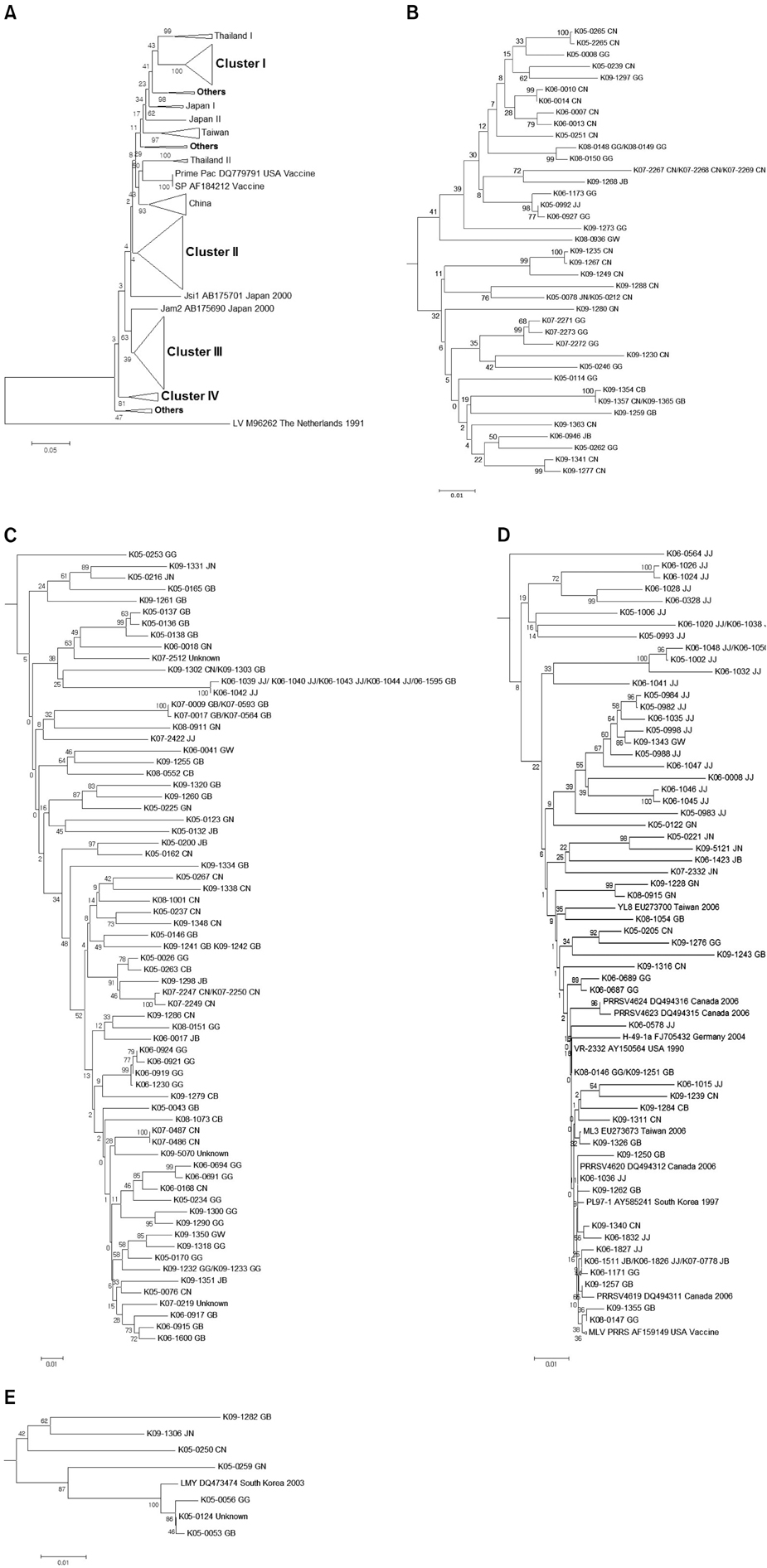
- The high genetic diversity of porcine reproductive and respiratory syndrome virus (PRRSV) has been an obstacle to developing an effective vaccine for porcine reproductive and respiratory syndrome (PRRS). This study was performed to assess the degree of genetic diversity among PRRSVs from Korean pig farms where wasting and respiratory syndrome was observed from 2005 to 2009. Samples from 786 farms were tested for the presence of PRRSV using reverse transcription PCR protocol. A total of 117 farms were positive for type 1 PRRSV while 198 farms were positive for type 2. Nucleotide sequences encoding the open reading frame (ORF) 5 were analyzed and compared to those of various published PRRSV isolates obtained worldwide. Sequence identity of the ORF 5 in the isolates was 81.6~100% for type 1 viruses and 81.4~100% for type 2 viruses. Phylogenetic analysis of the ORF 5 sequences showed that types 1 and 2 PRRSVs from Korea were mainly classified into three and four clusters, respectively. The analyzed isolates were distributed throughout the clusters independent of the isolation year or geographical origin. In conclusion, our results indicated that the genetic diversity of PRRSVs from Korean pig farms is high and has been increasing over time.
Keyword
MeSH Terms
-
Animal Husbandry
Animals
*Genes, Viral
*Genetic Variation
Lung/virology
Lymph Nodes/virology
*Open Reading Frames
Phylogeny
Porcine Reproductive and Respiratory Syndrome/virology
Porcine respiratory and reproductive syndrome virus/chemistry/classification/*genetics/isolation & purification
Republic of Korea
Reverse Transcriptase Polymerase Chain Reaction/veterinary
Sequence Analysis, DNA/veterinary
Sequence Analysis, Protein/veterinary
Swine
Figure
Cited by 2 articles
-
Survey of porcine respiratory disease complex-associated pathogens among commercial pig farms in Korea via oral fluid method
Yeotaek Cheong, Changin Oh, Kunkyu Lee, Ki-hyun Cho
J Vet Sci. 2017;18(3):283-289. doi: 10.4142/jvs.2017.18.3.283.An enhanced immunochromatographic strip test using colloidal gold nanoparticle-labeled dual-type N proteins for detection of antibodies to PRRS virus
Ji Eun Yu, In-Ohk Ouh, Hyeonjeong Kang, Hye-young Lee, Kwang-Myun Cheong, In-Soo Cho, Sang-Ho Cha
J Vet Sci. 2018;19(4):519-527. doi: 10.4142/jvs.2018.19.4.519.
Reference
-
1. Albina E. Epidemiology of porcine reproductive and respiratory syndrome (PRRS): an overview. Vet Microbiol. 1997; 55:309–316.
Article2. Allende R, Lewis TL, Lu Z, Rock DL, Kutish GF, Ali A, Doster AR, Osorio FA. North American and European porcine reproductive and respiratory syndrome viruses differ in non-structural protein coding regions. J Gen Virol. 1999; 80:307–315.
Article3. An TQ, Zhou YJ, Qiu HJ, Tong GZ, Wang YF, Liu JX, Yang JY. Identification of a novel B cell epitope on the nucleocapsid protein of porcine reproductive and respiratory syndrome virus by phage display. Virus Genes. 2005; 31:81–87.
Article4. Ansari IH, Kwon B, Osorio FA, Pattnaik AK. Influence of N-linked glycosylation of porcine reproductive and respiratory syndrome virus GP5 on virus infectivity, antigenicity, and ability to induce neutralizing antibodies. J Virol. 2006; 80:3994–4004.
Article5. Beyer J, Fichtner D, Schirrmeier H, Polster U, Weiland E, Wege H. Porcine reproductive and respiratory syndrome virus (PRRSV): kinetics of infection in lymphatic organs and lung. J Vet Med B Infect Dis Vet Public Health. 2000; 47:9–25.
Article6. Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997; 142:629–633.7. Cha SH, Choi EJ, Park JH, Yoon SR, Song JY, Kwon JH, Song HJ, Yoon KJ. Molecular characterization of recent Korean porcine reproductive and respiratory syndrome (PRRS) viruses and comparison to other Asian PRRS viruses. Vet Microbiol. 2006; 117:248–257.
Article8. Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003; 31:3497–3500.
Article9. Cho JG, Dee SA. Porcine reproductive and respiratory syndrome virus. Theriogenology. 2006; 66:655–662.
Article10. Choi EJ, Lee CH, Song JY, Song HJ. Survey on porcine reproductive and respiratory disease virus infection in pig farms associated with wasting and respiratory syndrome during 2005~2009 in Korea. Korean J Vet Public Health. 2011; 35:29–33.11. Dea S, Gagnon CA, Mardassi H, Pirzadeh B, Rogan D. Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the North American and European isolates. Arch Virol. 2000; 145:659–688.
Article12. Faaberg KS, Hocker JD, Erdman MM, Harris DLH, Nelson EA, Torremorell M, Plagemann PGW. Neutralizing antibody responses of pigs infected with natural GP5 N-glycan mutants of porcine reproductive and respiratory syndrome virus. Viral Immunol. 2006; 19:294–304.
Article13. Fang L, Jiang Y, Xiao S, Niu C, Zhang H, Chen H. Enhanced immunogenicity of the modified GP5 of porcine reproductive and respiratory syndrome virus. Virus Genes. 2006; 32:5–11.
Article14. Kim J, Chung HK, Chae C. Association of porcine circovirus 2 with porcine respiratory disease complex. Vet J. 2003; 166:251–256.
Article15. Kim J, Chung HK, Jung T, Cho WS, Choi C, Chae C. Postweaning multisystemic wasting syndrome of pigs in Korea: prevalence, microscopic lesions and coexisting microorganisms. J Vet Med Sci. 2002; 64:57–62.
Article16. Kim JY, Lee SY, Sur JH, Lyoo YS. Serological and genetic characterization of the European strain of the porcine reproductive and respiratory syndrome virus isolated in Korea. Korean J Vet Res. 2006; 46:363–370.17. Kim SH, Roh IS, Choi EJ, Lee C, Lee CH, Lee KH, Lee KK, Song YK, Lee OS, Park CK. A molecular analysis of European porcine reproductive and respiratory syndrome virus isolated in South Korea. Vet Microbiol. 2010; 143:394–400.
Article18. Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980; 16:111–120.
Article19. Labarque G, Van Reeth K, Nauwynck H, Drexler C, Van Gucht S, Pensaert M. Impact of genetic diversity of European-type porcine reproductive and respiratory syndrome virus strains on vaccine efficacy. Vaccine. 2004; 22:4183–4190.
Article20. Lee C, Kim H, Kang B, Yeom M, Han S, Moon H, Park S, Kim H, Song D, Park B. Prevalence and phylogenetic analysis of the isolated type I porcine reproductive and respiratory syndrome virus from 2007 to 2008 in Korea. Virus Genes. 2010; 40:225–230.
Article21. Mateu E, Diaz I. The challenge of PRRS immunology. Vet J. 2008; 177:345–351.
Article22. Meng XJ. Heterogeneity of porcine reproductive and respiratory syndrome virus: implications for current vaccine efficacy and future vaccine development. Vet Microbiol. 2000; 74:309–329.
Article23. Nelsen CJ, Murtaugh MP, Faaberg KS. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J Virol. 1999; 73:270–280.
Article24. Plagemann PGW. The primary GP5 neutralization epitope of North American isolates of porcine reproductive and respiratory syndrome virus. Vet Immunol Immunopathol. 2004; 102:263–275.
Article25. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987; 4:406–425.26. Snijder EJ, Meulenberg JJM. The molecular biology of arteriviruses. J Gen Virol. 1998; 79:961–979.
Article27. Stadejek T, Oleksiewicz MB, Scherbakov AV, Timina AM, Krabbe JS, Chabros K, Potapchuk D. Definition of subtypes in the European genotype of porcine reproductive and respiratory syndrome virus: nucleocapsid characteristics and geographical distribution in Europe. Arch Virol. 2008; 153:1479–1488.
Article28. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28:2731–2739.
Article29. Wu WH, Fang Y, Farwell R, Steffen-Bien M, Rowland RRR, Christopher-Hennings J, Nelson EA. A 10-kDa structural protein of porcine reproductive and respiratory syndrome virus encoded by ORF2b. Virology. 2001; 287:183–191.
Article30. Yoon SH, Song JY, Lee CH, Choi EJ, Cho IS, Kim B. Genetic characterization of the Korean porcine reproductive and respiratory syndrome viruses based on the nucleocapsid protein gene (ORF7) sequences. Arch Virol. 2008; 153:627–635.
Article31. Zheng Q, Chen D, Li P, Bi Z, Cao R, Zhou B, Chen P. Co-expressing GP5 and M proteins under different promoters in recombinant modified vaccinia virus ankara (rMVA)-based vaccine vector enhanced the humoral and cellular immune responses of porcine reproductive and respiratory syndrome virus (PRRSV). Virus Genes. 2007; 35:585–595.
Article32. Zhou YJ, Yu H, Tian ZJ, Li GX, Hao XF, Yan LP, Peng JM, An TQ, Xu AT, Wang YX, Wei TC, Zhang SR, Cai XH, Feng L, Li X, Zhang GH, Zhou LJ, Tong GZ. Genetic diversity of the ORF5 gene of porcine reproductive and respiratory syndrome virus isolates in China from 2006 to 2008. Virus Res. 2009; 144:136–144.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Genetic diversity and phylogenetic analysis of porcine reproductive and respiratory syndrome virus in southern China from 2007 to 2014
- Intracellular Localization of the Porcine Reproductive and Respiratory Syndrome Virus Nucleocapsid Protein
- Porcine ear necrosis syndrome by coinfection of porcine reproductive and respiratory syndrome virus and Staphylococcus hyicus
- Prevalence of porcine reproductive and respiratory syndrome virus, porcine circovirus type 2 and porcine parvovirus from aborted fetuses and pigs with respiratory problems in Korea
- Phylogenetic characterization of genes encoding for glycoprotein 5 and membrane protein of PRRSV isolate HH08