J Bacteriol Virol.
2007 Sep;37(3):153-160. 10.4167/jbv.2007.37.3.153.
Utility of RT-PCR-based Dot-blot Hybridization for Detecting and Genotyping Echoviruses
- Affiliations
-
- 1National Institute of Health, Korea Centers for Disease Control and Prevention, Seoul 122-701, Republic of Korea. jooshil@nih.go.kr
- KMID: 1513570
- DOI: http://doi.org/10.4167/jbv.2007.37.3.153
Abstract
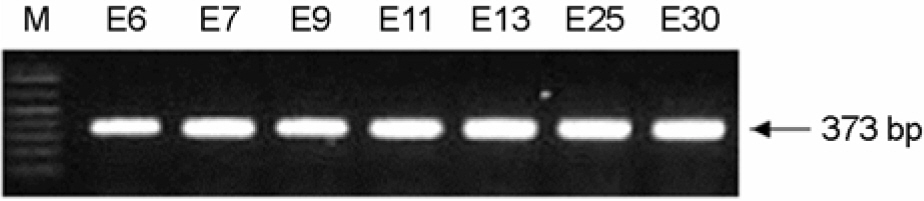
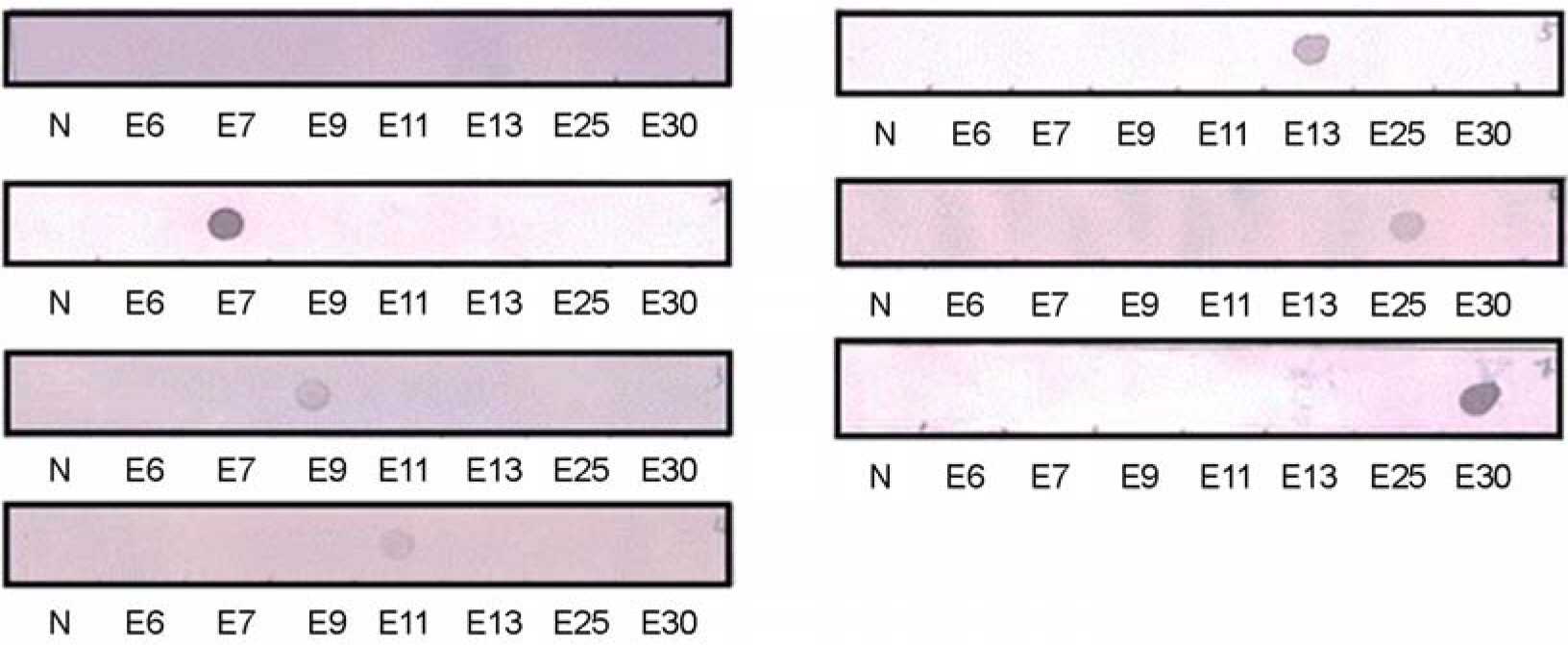
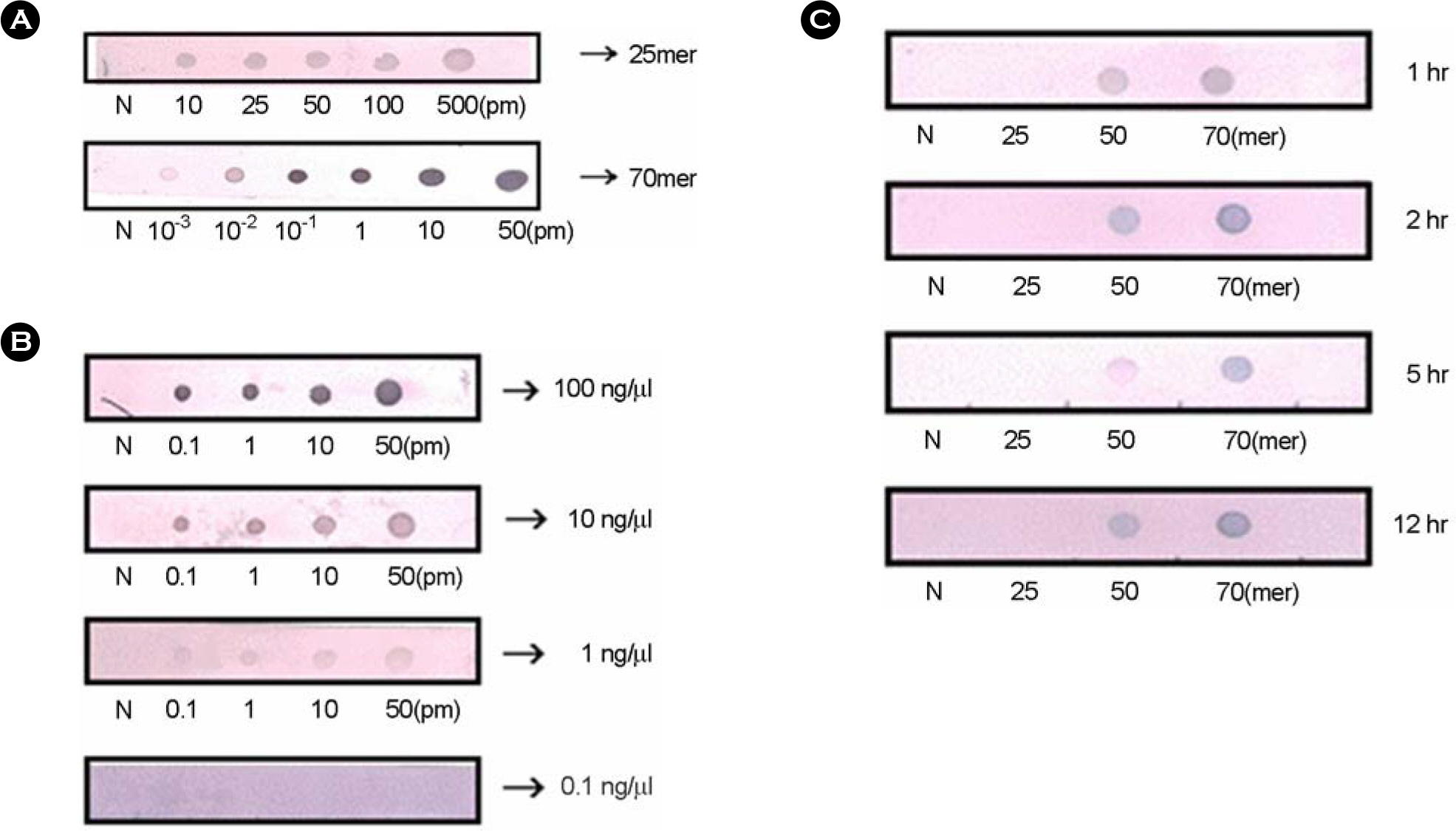
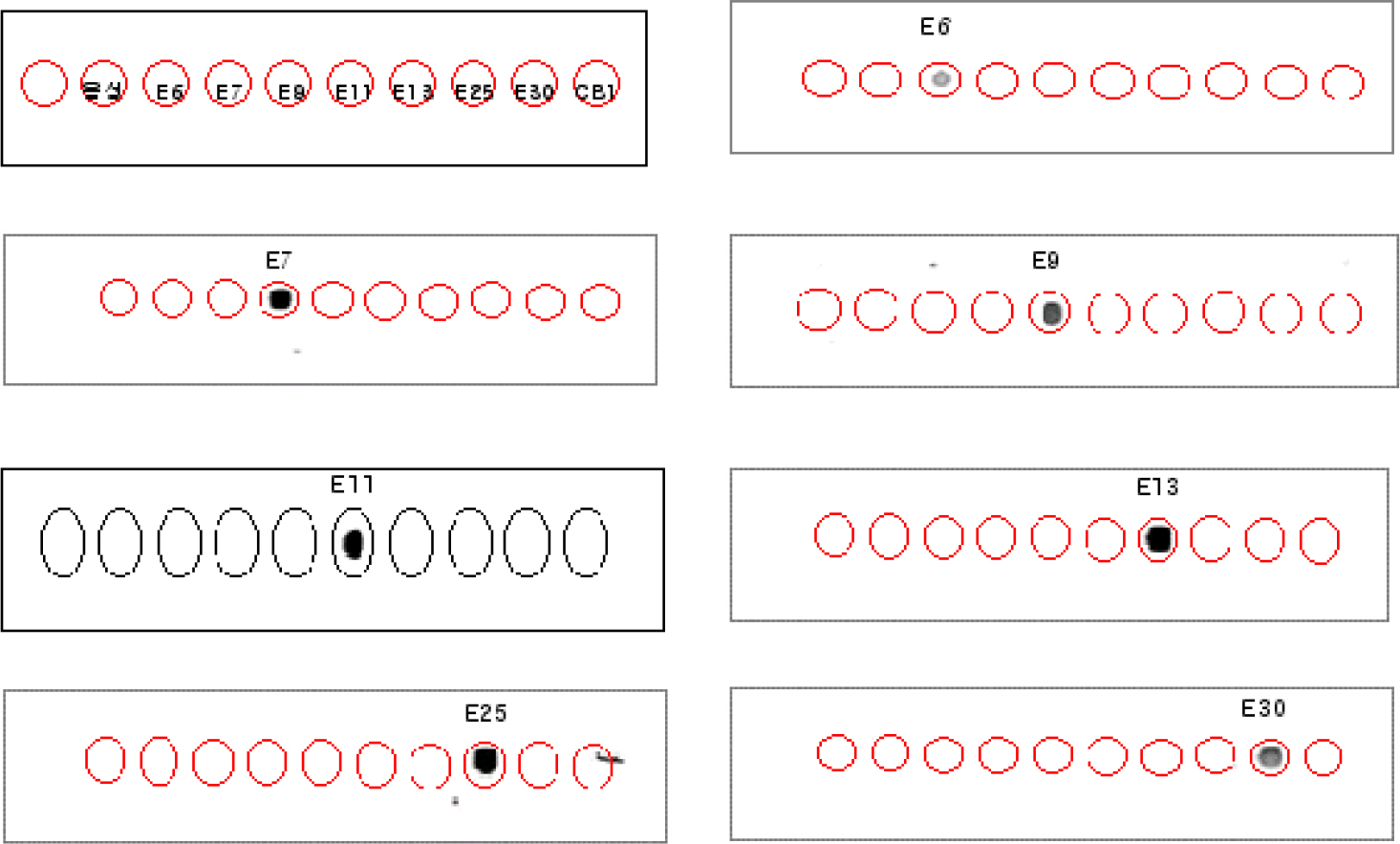
- We attempted to detect and identify virus types quickly by improving an RT-PCR-based dot-blot hybridization test for echoviruses, important human pathogens mainly causing aseptic meningitis. This test was applied to reference viruses of seven echovirus serotypes prevalent in Korea (E6, 7, 9, 11, 13, 25, and 30) and seventy isolates of echovirus isolated in Korea between 2002 and 2004. The primers for target DNA and hybridization probes (25mer, 50mer, and 70mer) were designed within the VP1 region of the echovirus. In RT-PCR, a nonradioactive digoxigenin-DNA labeling mix was added instead of dNTP to initiate PCR. The PCR product was then hybridized against 25mer, 50mer, and 70mer probe DNA spotted on nylon membranes and the reaction was observed. To investigate the optimal conditions for hybridization, various concentrations of target DNA (0.1, 1, 10, and 100 ng/micron l), size of probe DNA (25mer, 50mer, and 70mer), concentrations of probe DNA (10~50 pM), and reaction time were included. In the test zone, the optimal condition in terms of time and cost was a reaction time of 1 h with 10 ng/micron l target DNA concentration and 10 pM of a 50mer probe. We found 100% diagnosis of the serotypes for seven reference echoviruses and 90% (63/70) sensitivity for clinical isolates. Also, tests with this probe for reactivity with seven reference echoviruses by using DNA chips showed that diagnostic identification was possible without other serotype cross-reactivity. Therefore, efficiency analysis of probe and target DNA on clinical specimens by using dot-blot analysis indicated that this system can be applied to the prestages of the DNA chip and that the dot blot analysis itself can be used in applications to develop a tool for diagnosing specific viral serotypes.
Keyword
MeSH Terms
Figure
Reference
-
References
1). Arola A, Santti J, Ruuskanen O, Halonen P, Hyypia T. Identification of enteroviruses in clinical specimens by competitive PCR followed by genetic typing using sequence analysis. J Clin Microbiol. 34:313–318. 1996.
Article2). Bottner A, Daneschnejad S, Handrick W, Schuster V, Liebert UG, Kiess W. A season of aseptic meningitis in Germany: epidemiologic, clinical and diagnostic aspects. Pediatr Infect Dis J. 21:112632. 2002.3). Caro V, Guillot S, Delpeyroux F, Crainic R. Molecular strategy for ‘serotyping’ of human enteroviruses. J Gen Virol. 82:79–91. 2001.
Article4). Chomel JJ, Antona D, Thouvenot D, Lina B. Three ECHOvirus serotypes responsible for outbreak of aseptic meningitis in Rhone-Alpes region, France. Eur J Clin Microbiol Infect Dis. 22:191–193. 2003.5). Jee YM, Cheon DS, Choi WY, Ahn JB, Kim KS, Chung YS, Lee JW, Lee KB, Noh HS, Park KS, Lee SH, Kim SH, Cho KS, Kim ES, Jung JK, Yoon JD, Cho HW. Updates on enterovirus surveillance in Korea: 1999–2003. Infection and Chemotherapy. 36:294–303. 2004.6). Kirschke DL, Jones TF, Buckingham SC, Craig AS, Schaffner W. Outbreak of aseptic meningitis associated with echovirus 13. Pediatr Infect Dis J. 21(11):1034–1038. 2002.
Article7). Kottaridi C, Bolanaki E, Siafakas N, Markoulatos P. Evaluation of seroneutralization and molecular diagnostic methods for echovirus identification. Diagn Microbiol Infect Dis. 53:113–119. 2005.
Article8). Lim KA, Benyesh-Melnick M. Typing of viruses by combinations of antiserum pools. Application to typing of enteroviruses (Coxsackie and ECHO). J Immunol. 84:309–317. 1960.9). Lin TY, Twu SJ, Ho MS, Chang LY, Lee CY. Entero-virus 71 outbreaks, Taiwan: occurrence and recognition. Emerg Infect Dis. 9:291–293. 2003.10). Mateu MG. Antibody recognition of picornaviruses and escape from neutralization: a structural view. Virus Res. 38:1–24. 1995.
Article11). Melnick JL. Enteroviruses: polioviruses, coxsackie-viruses, echoviruses and newer enteroviruses. pp. p. 655–712. In:. Fields Virology. 3rd ed.Fields BN, Knipe DM, Howley PM, editors. (Ed),. Lipincott-Raven;Philadelphia: 1996.12). Melnick JL, Rennick V, Hampil B, Schmidt NJ, Ho HH. Lyophilized combination pools of enterovirus equine antisera: preparation and test procedures for the identification of field strains of 42 enteroviruses. Bull World Health Organ. 48:263–268. 1973.13). Minor PD. Antigenic structure of picornaviruses. Curr Top Microbiol Immunol. 161:121–154. 1990.
Article14). Oberste MS, Maher K, Kilpatrick DR, Pallansch MA. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol. 73:1941–1948. 1999.
Article15). Oberste MS, Nix WA, Maher K, Pallansch MA. Improved molecular identification of enteroviruses by RT-PCR and amplicon sequencing. J Clin Virol. 26:375–377. 2003.
Article16). Takami T, Nakayama T, Kawashima H, Takei Y, Takekuma K, Hoshika A. Determination of entero-virus serotype inferred from sequence analysis of PCR products. J Clin Virol. 26:355–359. 2003.
Article17). Thoelen I, Lemey P, Van Der Donck I, Beuselinck K, Lindberg AM, Van Ranst M. Molecular typing and epidemiology of enteroviruses identified from an outbreak of aseptic meningitis in Belgium during the summer of 2000. J Med Virol. 70(3):420–429. 2003.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Sensitive and Specific PCR/dot Blot Hybridization assay for the Detection of Ovine Herpesvirus 2, a Gamma Herpesvirus
- Sensitive and Specific Detection of Mycoplasma species by Consensus Polymerase Chain Reaction and Dot Blot Hybridization
- Genotyping of the Platelet Alloantigens by Reverse Dot Blot Hybridization
- Development of Streptococcus sanguinis-, Streptococcus parasanguinis-, and Streptococcus gordonii-PCR Primers Based on the Nucleotide Sequences of Species-specific DNA Probes Screened by Inverted Dot Blot Hybridization
- Detection of canine respiratory coronavirus from dogs with respiratory disease