J Bacteriol Virol.
2008 Dec;38(4):259-266. 10.4167/jbv.2008.38.4.259.
Development of UPSR Primer Design Program for Efficient Detection of Viruses
- Affiliations
-
- 1Department of Microbiology, Chungbuk National University, Cheongju, Chungbuk, Korea. chlee@cbu.ac.kr
- 2Department of Clinical Pathology, Juseong College, Nesu-eup, Chungbuk, Korea.
- 3Department of Bioinformation Technique, Chungbuk National University, Cheongju, Chungbuk, Korea.
- 4School of Computer Science and Engineering, Chungbuk National University, Cheongju, Chungbuk, Korea.
- KMID: 1483967
- DOI: http://doi.org/10.4167/jbv.2008.38.4.259
Abstract
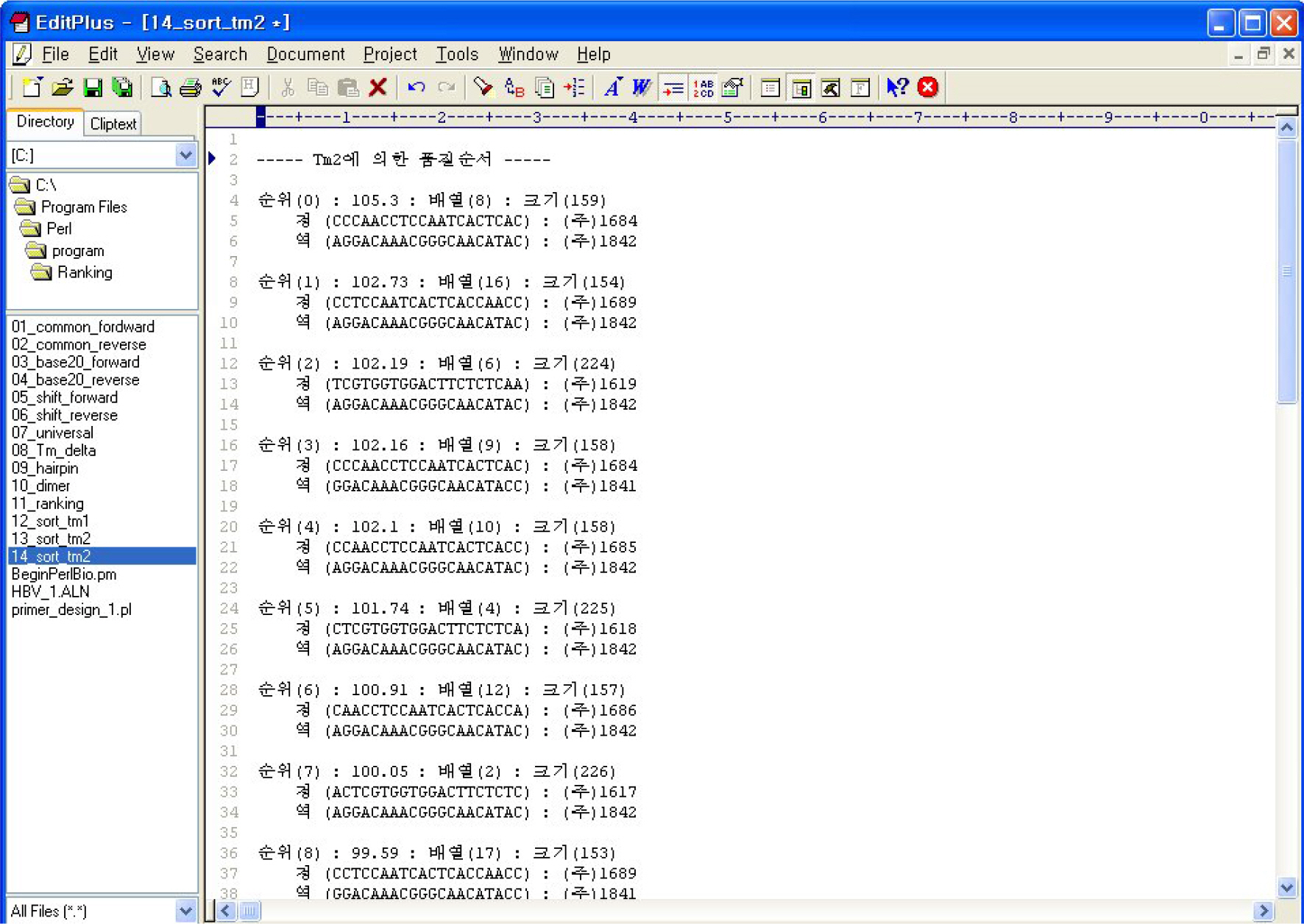
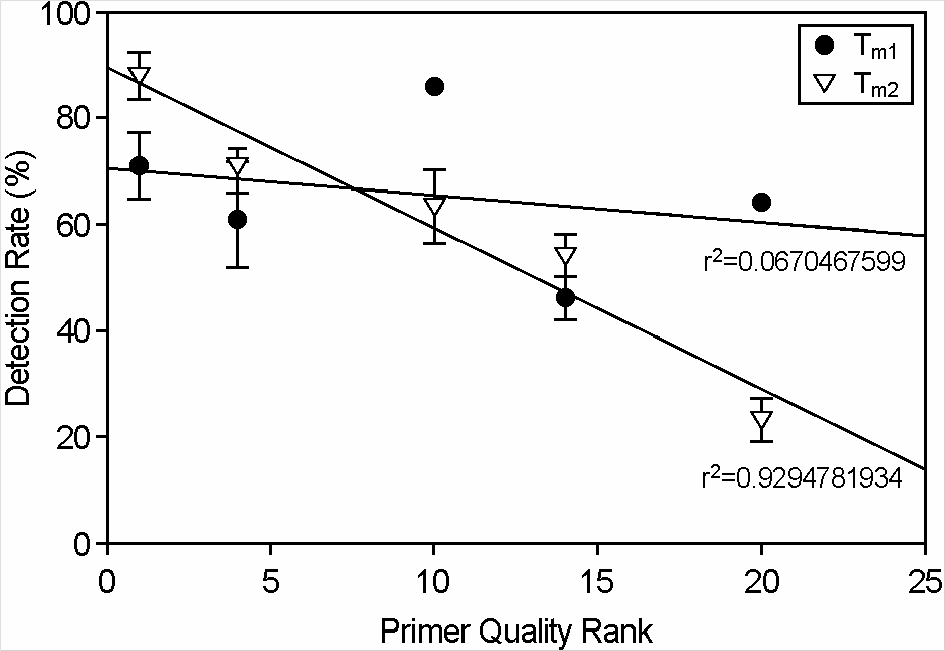
- PCR is a rapid and sensitive method for detection of viruses from clinical samples and good primers are essential for successful PCR. However, high mutation rate of viral genomes often results in failure in detecting viruses, and there have been attempts to develop primers from multiple viral sequences. Thus, we developed a program called Universal Primers Score Ranking (UPSR) which generates primers from multiple sequences and ranks the quality of primers automatically. The feasibility of the UPSR program was tested using hepatitis B viruses (HBV) isolated from Korean patients. UPSR generated primer candidates with quality score ranks according to two T(m) values. We found that T(m2) values calculated based on the thermodynamics of nearest neighboring bases were better correlated with actual detection rate of HBV from patients' sera. The primer with number 1 rank by T(m2) values detected more samples than any other primers designed by UPSR, commercial primer, or other reference primers suggested by previous literatures. Thus, UPSR proved to be easy and useful to design primers from multiples sequences in detecting viruses.
Keyword
MeSH Terms
Figure
Reference
-
1). Breslauer KJ., Frank R., Blocker H., Marky LA. Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci USA. 83:3746–3750. 1986.
Article2). Chen SH., Lin CY., Cho CS., Lo CZ., Hsiung CA. Primer Design Assistant (PDA): A web-based primer design tool. Nucleic Acids Res. 31:3751–3754. 2003.
Article3). Grant PR., Busch MP. Nucleic acid amplification technology methods used in blood donor screening. Transfus Med. 12:229–242. 2002.
Article4). Haas S., Vingron M., Poustka A., Wiemann S. Primer design for large scale sequencing. Nucleic Acids Res. 26:3006–3012. 1998.
Article5). Hamasaki K., Nakata K., Nagayama Y., Ohtsuru A., Daikoku M., Taniguchi K., Tsutsumi T., Sato Y., Kato Y., Nagataki S. Changes in the prevalence of HBeAg-negative mutant hepatitis B virus during the course of chronic hepatitis B. Hepatology. 20:8–14. 1994.
Article6). Huang YC., Chang CF., Chan CH., Yeh TJ., Chang YC., Chen CC., Kao CY. Integrated minimum-set primers and unique probe design algorithms for differential detection on symptom-related pathogens. Bioinformatics. 21:4330–4337. 2005.
Article7). Jarman SN. Amplicon: software for designing PCR primers on aligned DNA sequences. Bioinformatics. 20:1644–1645. 2004.
Article8). Koressaar T., Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 23:1289–1291. 2007.
Article9). Li P., Kupfer KC., Davies CJ., Burbee D., Evans GA., Garner HR. PRIMO: a primer design program that applies base quality statistics for automated large-scale DNA sequencing. Genomics. 40:476–485. 1997.
Article10). Nainan OV., Cromeans TL., Margolis HS. Sequence-specific, single-primer amplification and detection of PCR products for identification of hepatitis viruses. J Virol Methods. 61:127–134. 1996.
Article11). Proutski V., Holmes EC. Primer Master: a new program for the design and analysis of PCR primers. Comput Appl Biosci. 12:253–255. 1996.
Article12). Raddatz G., Dehio M., Meyer TF., Dehio C. PrimeArray: genome-scale primer design for DNA-microarray construction. Bioinformatics. 17:98–99. 2001.
Article13). Roth WK., Weber M., Petersen D., Drosten C., Buhr S., Sireis W., Weichert W., Hedges D., Seifried E. NAT for HBV and anti-HBc testing increase blood safety. Transfusion. 42:869–875. 2002.14). Rozen S., Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. pp. p. 365–386. In. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Misener S, Krawetz SA, editors. (Ed),. Humana Press;Totowa, NJ: 2000.
Article15). SantaLucia J Jr., Allawi HT., Seneviratne PA. Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry. 35:3555–3562. 1996.
Article16). Sugimoto N., Nakano S., Yoneyama M., Honda K. Improved thermodynamic parameters and helix initiation factor to predict stability of DNA duplexes. Nucleic Acids Res. 24:4501–4505. 1996.
Article17). Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen JA. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35:W71–74. 2007.
Article18). Wang D., Urisman A., Liu YT., Springer M., Ksiazek TG., Erdman DD., Mardis ER., Hickenbotham M., Magrini V., Eldred J., Latreille JP., Wilson RK., Ganem D., DeRisi JL. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 1:E2. 2003.
Article19). Zaaijer HL., ter Borg F., Cuypers HT., Hermus MC., Lelie PN. Comparison of methods for detection of hepatitis B virus DNA. J Clin Microbiol. 32:2088–2091. 1994.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Development and Verification of Nested PCR Assay for Detection of Tobacco rattle virus in Plant Quarantine
- PCR Assays for Detection of Pseudomonas tolaasii and Pseudomonas agarici
- Comparison of Culture, Direct Immunofluorescence Assay, and Multiplex Reverse Transcriptase PCR for Detection of Respiratory Viruses
- Development of Efficient Screening Methods for Hazardous HBV Virus in Vaccine Preparation
- PCR-Based Sensitive Detection of Wood-Decaying Fungus Phellinus linteus by Specific Primer from rDNA ITS Regions