Healthc Inform Res.
2024 Oct;30(4):355-363. 10.4258/hir.2024.30.4.355.
Mapping Drug Terms via Integration of a Retrieval-Augmented Generation Algorithm with a Large Language Model
- Affiliations
-
- 1Department of Medical Informatics, Medical School of Ehime University, Toon, Ehime, Japan
- 2Yuimedi Inc., Tokyo, Japan
- KMID: 2560538
- DOI: http://doi.org/10.4258/hir.2024.30.4.355
Abstract
Objectives
This study evaluated the efficacy of integrating a retrieval-augmented generation (RAG) model and a large language model (LLM) to improve the accuracy of drug name mapping across international vocabularies.
Methods
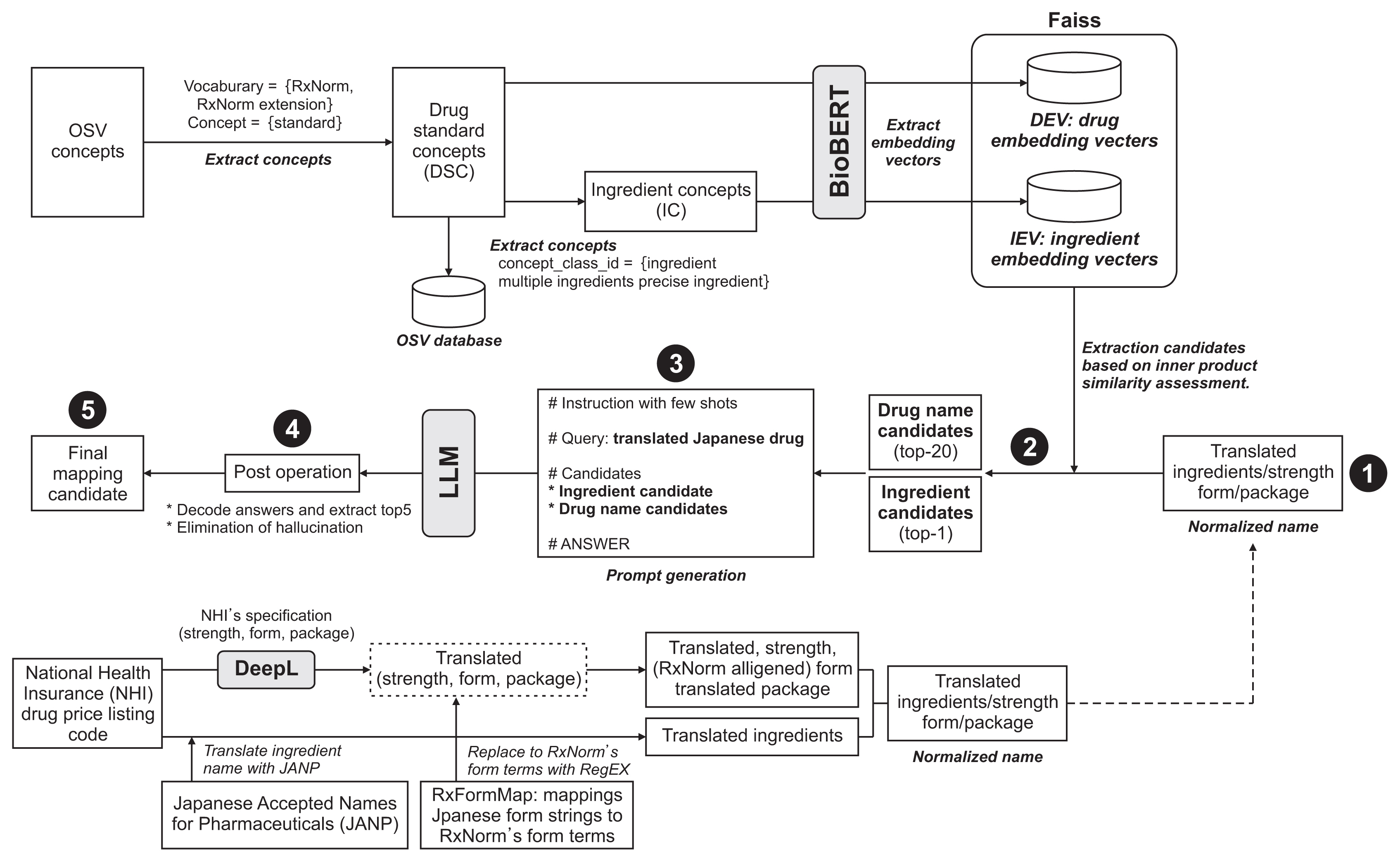
Drug ingredient names were translated into English using the Japanese Accepted Names for Pharmaceuticals. Drug concepts were extracted from the standard vocabulary of OHDSI, and the accuracy of mappings between translated terms and RxNorm was assessed by vector similarity, using the BioBERT-generated embedded vectors as the baseline. Subsequently, we developed LLMs with RAG that distinguished the final candidates from the baseline. We assessed the efficacy of the LLM with RAG in candidate selection by comparing it with conventional methods based on vector similarity.
Results
The evaluation metrics demonstrated the superior performance of the combined LLM + RAG over traditional vector similarity methods. Notably, the hit rates of the Mixtral 8x7b and GPT-3.5 models exceeded 90%, significantly outperforming the baseline rate of 64% across stratified groups of PO drugs, injections, and all interventions. Furthermore, the r-precision metric, which measures the alignment between model judgment and human evaluation, revealed a notable improvement in LLM performance, ranging from 41% to 50% compared to the baseline of 23%.
Conclusions
Integrating an RAG and an LLM outperformed conventional string comparison and embedding vector similarity techniques, offering a more refined approach to global drug information mapping.
Figure
Reference
-
References
1. Hripcsak G, Duke JD, Shah NH, Reich CG, Huser V, Schuemie MJ, et al. Observational Health Data Sciences and Informatics (OHDSI): opportunities for observational researchers. Stud Health Technol Inform. 2015; 216:574–8.2. Stang PE, Ryan PB, Racoosin JA, Overhage JM, Hartzema AG, Reich C, et al. Advancing the science for active surveillance: rationale and design for the observational medical outcomes partnership. Ann Intern Med. 2010; 153(9):600–6. https://doi.org/10.7326/0003-4819-153-9-201011020-00010.
Article3. Reich C, Ostropolets A, Ryan P, Rijnbeek P, Schuemie M, Davydov A, et al. OHDSI standardized vocabularies-a large-scale centralized reference ontology for international data harmonization. J Am Med Inform Assoc. 2024; 31(3):583–90. https://doi.org/10.1093/jamia/ocad247.
Article4. Wang L, Zhang Y, Jiang M, Wang J, Dong J, Liu Y, et al. Toward a normalized clinical drug knowledge base in China-applying the RxNorm model to Chinese clinical drugs. J Am Med Inform Assoc. 2018; 25(7):809–18. https://doi.org/10.1093/jamia/ocy020.
Article5. Henke E, Zoch M, Kallfelz M, Ruhnke T, Leutner LA, Spoden M, et al. Assessing the use of German claims data vocabularies for research in the observational medical outcomes partnership common data model: development and evaluation study. JMIR Med Inform. 2023; 11:e47959. https://doi.org/10.2196/47959.
Article6. Maier C, Lang L, Storf H, Vormstein P, Bieber R, Bernarding J, et al. Towards implementation of OMOP in a German University Hospital Consortium. Appl Clin Inform. 2018; 9(1):54–61. https://doi.org/10.1055/s-0037-1617452.
Article7. Nelson SJ, Zeng K, Kilbourne J, Powell T, Moore R. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc. 2011; 18(4):441–8. https://doi.org/10.1136/amiajnl-2011-000116.
Article8. de Groot R, Puttmann DP, Fleuren LM, Thoral PJ, Elbers PW, de Keizer NF, et al. Determining and assessing characteristics of data element names impacting the performance of annotation using Usagi. Int J Med Inform. 2023. Oct. 178:105200. https://doi.org/10.1016/j.ijmedinf.2023.105200.
Article9. Zhang Y, Jin R, Zhou ZH. Understanding bag-of-words model: a statistical framework. Int J Mach Learn Cybern. 2010; 1(1):43–52. https://doi.org/10.1007/s13042-010-0001-0.
Article10. Butler J, Zand M. Similarity mapping of national drug code formulary systems between nations [Internet]. Durham (NC): Research Square;2022. [cited at 2024 Jun 6]. Available from: https://doi.org/10.21203/rs.3.rs-1858694/v1.
Article11. Zhang Y, Guo L, Du C, Wang Y, Huang D. Extraction of English drug names based on Bert-CNN mode. J Inf Hiding Multimed Signal Process. 2020; 11(2):70–8.12. Brown T, Mann B, Ryder N, Subbiah M, Kaplan JD, Dhariwal P, et al. Language models are few-shot learners. Adv Neural Inf Process Syst. 2020; 33:1877–901. https://doi.org/10.48550/arXiv.2005.14165.
Article13. Gao Y, Xiong Y, Gao X, Jia K, Pan J, Bi Y, et al. Retrieval-augmented generation for large language models: a survey [Internet]. Ithaca (NY): arXiv.org;2023. [cited at 2024 Jun 6]. Available from: https://arxiv.org/abs/2005.14165.14. Izacard G, Lewis P, Lomeli M, Hosseini L, Petroni F, Schick T, et al. Few-shot learning with retrieval augmented language models [Internet]. Ithaca (NY): arXiv.org;2022. [cited at 2024 Jun 6]. Available from: https://arxiv.org/abs/2208.03299.15. Ministry of Health, Labour and Welfare. National Health Insurance (NHI) drug price listing code 2024 [Internet]. Tokyo, Japan: Ministry of Health, Labour and Welfare;c2024. [cited at 2024 Jun 6]. Available from: https://www.mhlw.go.jp/stf/seisakunitsuite/bunya/0000078916.html.16. National Institute of Health Sciences. Japanese accepted names for pharmaceuticals [Internet]. Tokyo, Japan: National Institute of Health Sciences;c2024. [cited at 2024 Jun 6]. Available from: https://jpdb.nihs.go.jp/jan.17. Odysseus Data Services. ATHENA: OHDSI Vocabularies Repository [Internet]. [place unknown]: OHDSI;c2024. [cited at 2024 Jun 6]. Available from: https://athena.ohdsi.org/.18. Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, et al. BioB-ERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020; 36(4):1234–40. https://doi.org/10.1093/bioinformatics/btz682.
Article19. Wu Y. Google’s neural machine translation system: bridging the gap between human and machine translation [Internet]. Ithaca (NY): arXiv.org;2016. [cited at 2024 Jun 6]. Available from: https://arxiv.org/abs/1609.08144.20. Douze M, Guzhva A, Deng C, Johnson J, Szilvasy G, Mazare PE, et al. The Faiss library [Internet]. Ithaca (NY): arXiv.org;2024. [cited at 2024 Jun 6]. Available from: https://arxiv.org/abs/2401.08281.21. Lomeli M. Faiss indexes [Internet]. San Francisco (CA): GitHub;2023. [cited at 2024 Jun 6]. Available from: https://github.com/facebookresearch/faiss/wiki/Faissindexes.22. Jiang AQ, Sablayrolles A, Mensch A, Bamford C, Chaplot DS, de las Casas D, et al. Mistral 7B [Internet]. Ithaca (NY): arXiv.org;2023. [cited at 2024 Jun 6]. Available from: https://arxiv.org/abs/2310.06825.23. Tunstall L, Schmid P, Sanseviero O, Cuenca P, Dehaene O, von Werra L, et al. Welcome Mixtral: a SOTA mixture of experts on Hugging Face [Internet]. Brooklyn (NY): Hugging Face;2023. [cited at 2024 Jun 6]. Available from: https://huggingface.co/blog/mixtral.24. Ji Z, Wei Q, Xu H. BERT-based ranking for biomedical entity normalization. AMIA Jt Summits Transl Sci Proc. 2020; 2020:269–77.25. Shazeer N, Mirhoseini A, Maziarz K, Davis A, Le Q, Hinton G, et al. Outrageously large neural networks: the sparsely-gated mixture-of-experts layer [Internet]. Ithaca (NY): arXiv.org;2017. [cited at 2024 Jun 6]. Available from: https://arxiv.org/abs/1701.06538.26. Achiam J, Adler S, Agarwal S, Ahmad L, Akkaya I, Aleman FL, et al. GPT-4 technical report [Internet]. Ithaca (NY): arXiv.org;2023. [cited at 2024 Jun 6]. Available from: https://arxiv.org/abs/2303.08774v1.27. Wu T, He S, Liu J, Sun S, Liu K, Han QL, et al. A brief overview of ChatGPT: the history, status quo and potential future development. IEEE/CAA J Automatica Sinica. 2023; 10(5):1122–36. https://doi.org/10.1109/JAS.2023.123618.
Article28. GitHub. LangChain [Internet]. San Francisco (CA): GitHub;2022. [cited at 2024 Jun 6]. Available from: https://github.com/langchain-ai/langchain.29. Bassani E. ranx: a blazing-fast python library for ranking evaluation and comparison. European Conference on Information Retrieval. Cham, Switzerland: Springer;2022. p. 259–64. https://doi.org/10.1007/978-3-030-99739-7_30.
Article30. Jarvelin K, Kekalainen J. Cumulated gain-based evaluation of IR techniques. ACM Trans Inf Syst. 2002; 20(4):422–46. https://doi.org/10.1145/582415.582418.
Article31. Moffat A, Zobel J. Rank-biased precision for measurement of retrieval effectiveness. ACM Trans Inf Syst. 2008; 27(1):1–27. https://doi.org/10.1145/1416950.1416952.
Article32. Chen Y, Hu D, Li M, Duan H, Lu X. Automatic SNOMED CT coding of Chinese clinical terms via attention-based semantic matching. Int J Med Inform. 2022; 159:104676. https://doi.org/10.1016/j.ijmedinf.2021.104676.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Comparative Study on the 'Sign or Symptom' Concepts of the UMLS(Unified Medical Language System) and Clinical Terms in Korean Medical Records
- The Integration of Large Language Models Such as ChatGPT in Scientific Writing: Harnessing Potential and Addressing Pitfalls
- Effective Query Expansion using Condensed UMLS Metathesaurus for Medical Information Retrieval
- ChatGPT’s Potential in Dermatology Knowledge Retrieval: An Analysis Using Korean Dermatology Residency Examination Questions
- Proposed Algorithm with Standard Terminologies (SNOMED and CPT) for Automated Generation of Medical Bills for Laboratory Tests