Ann Lab Med.
2024 Sep;44(5):446-449. 10.3343/alm.2023.0444.
Multiple Primary Cancers With Hematologic Malignancies and Germline Predisposition: A Case Series
- Affiliations
-
- 1Department of Laboratory Medicine, Korea University Anam Hospital, Korea University College of Medicine, Seoul, Korea
- 2Department of Laboratory Medicine, Seoul National University College of Medicine, Seoul, Korea
- 3Department of Genomic Medicine, Seoul National University Hospital, Seoul, Korea
- KMID: 2559151
- DOI: http://doi.org/10.3343/alm.2023.0444
Abstract
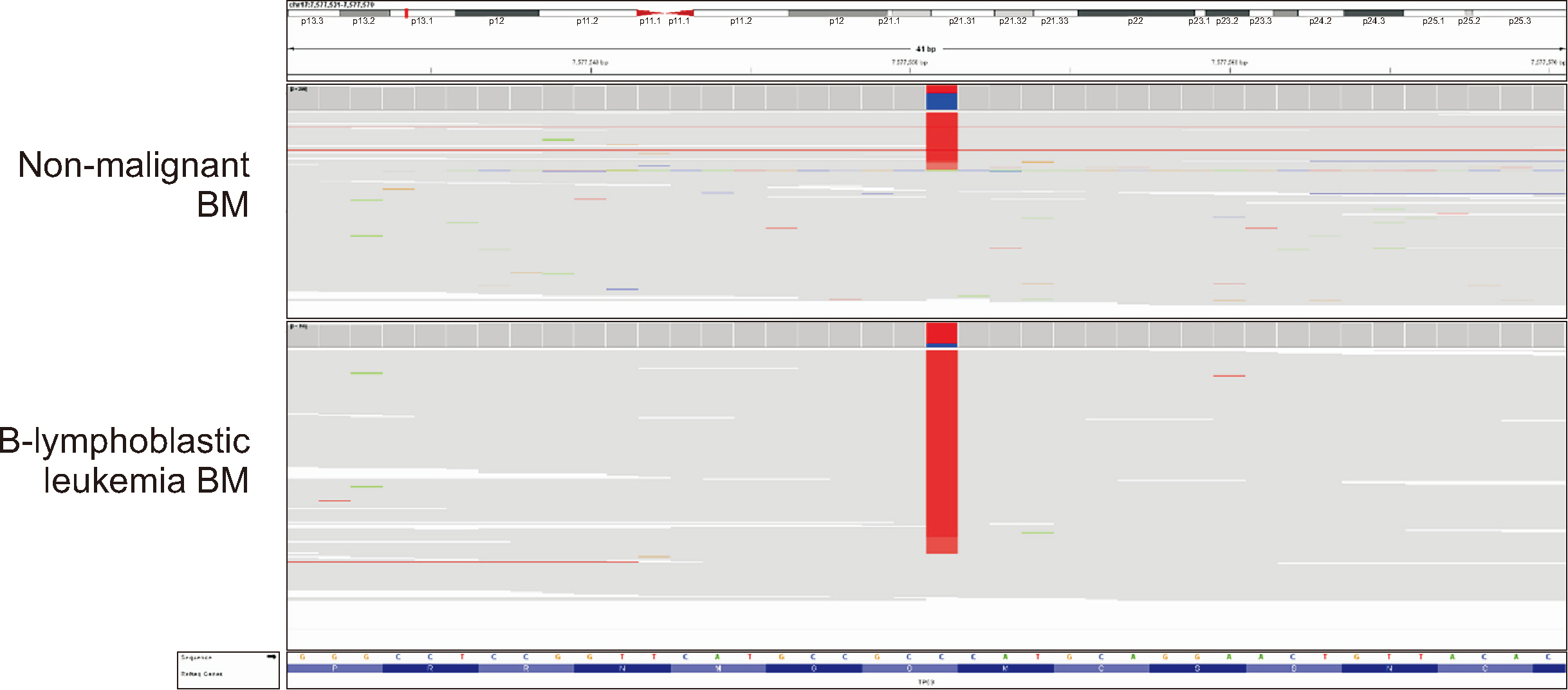
- The term “multiple primary (MP) cancers” refers to the existence of more than one cancer in the same patient. The combination of MP cancers with hematological malignancies is relatively uncommon. In this study, we present five patients diagnosed with MP cancers concomitant with hematological malignancies. We comprehensively analyzed their clinical characteristics, cytogenetic profiles, and germline and somatic variants. As first primaries, two patients had solid cancer not followed by cytotoxic therapy and three had hematologic cancer, followed by cytotoxic therapy. The second primaries were all hematologic malignancies that did not meet the criteria for therapy-related myeloid neoplasm. Notably, two (40%) out of the five patients harbored pathogenic potential/presumed germline variants in cancer predisposition genes. Therefore, germline variant testing should be considered when MP cancers with hematological malignancies require consideration for related donor stem cell transplantation.
Keyword
Figure
Reference
-
References
1. Vogt A, Schmid S, Heinimann K, Frick H, Herrmann C, Cerny T, et al. 2017; Multiple primary tumours: challenges and approaches, a review. ESMO Open. 2:e000172. DOI: 10.1136/esmoopen-2017-000172. PMID: 28761745. PMCID: PMC5519797.
Article2. Wang C, Shen Y, Zhang Y, Guo F, Li Q, Zhang H, et al. 2022; Metachronous multiple primary carcinoma with acute promyelocytic leukemia: 2 cases report and literature review. Front Oncol. 12:893319. DOI: 10.3389/fonc.2022.893319. PMID: 35756676. PMCID: PMC9214198. PMID: b745d4ed071942389aafdbf624181f94.
Article3. Adamo M, Dickie L, et al. 2014. SEER program coding and staging manual 2014. National Cancer Institute;Bethesda:4. International Association of Cancer Registries. 2005; International rules for multiple primary cancers. Asian Pac J Cancer Prev. 6:104–6.5. Working Group Report. 2005; International rules for multiple primary cancers (ICD-0 third edition). Eur J Cancer Prev. 14:307–8. DOI: 10.1097/00008469-200508000-00002. PMID: 16030420.6. Coyte A, Morrison DS, McLoone P. 2014; Second primary cancer risk - the impact of applying different definitions of multiple primaries: results from a retrospective population-based cancer registry study. BMC Cancer. 14:272. DOI: 10.1186/1471-2407-14-272. PMID: 24742063. PMCID: PMC4005906.
Article7. Buiatti E, Crocetti E, Acciai S, Gafà L, Falcini F, Milandri C, et al. 1997; Incidence of second primary cancers in three Italian population-based cancer registries. Eur J Cancer. 33:1829–34. DOI: 10.1016/S0959-8049(97)00173-1. PMID: 9470841.
Article8. Weir HK, Johnson CJ, Thompson TD. 2013; The effect of multiple primary rules on population-based cancer survival. Cancer Causes Control. 24:1231–42. DOI: 10.1007/s10552-013-0203-3. PMID: 23558444. PMCID: PMC4558881.
Article9. Rosso S, De Angelis R, Ciccolallo L, Carrani E, Soerjomataram I, Grande E, et al. 2009; Multiple tumours in survival estimates. Eur J Cancer. 45:1080–94. DOI: 10.1016/j.ejca.2008.11.030. PMID: 19121933.
Article10. Whitworth J, Smith PS, Martin JE, West H, Luchetti A, Rodger F, et al. 2018; Comprehensive cancer-predisposition gene testing in an adult multiple primary tumor series shows a Broad Range of deleterious variants and atypical tumor phenotypes. Am J Hum Genet. 103:3–18. DOI: 10.1016/j.ajhg.2018.04.013. PMID: 29909963. PMCID: PMC6037202.11. Swerdlow SH, Campo E, editors. 2017. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th ed. International Agency for Research on Cancer;Lyon: DOI: 10.53347/rid-9250.12. WHO Classification of Tumours. 2022. Haematolymphoid tumours. 5th ed. International Agency for Research on Cancer;Lyon: https://tumourclassification.iarc.who.int/chapters/63. DOI: 10.1038/s41375-022-01625-x.13. Churpek JE, Marquez R, Neistadt B, Claussen K, Lee MK, Churpek MM, et al. 2016; Inherited mutations in cancer susceptibility genes are common among survivors of breast cancer who develop therapy-related leukemia. Cancer. 122:304–11. DOI: 10.1002/cncr.29615. PMID: 26641009. PMCID: PMC4707981.
Article14. Schulz E, Valentin A, Ulz P, Beham-Schmid C, Lind K, Rupp V, et al. 2012; Germline mutations in the DNA damage response genes BRCA1, BRCA2, BARD1 and TP53 in patients with therapy related myeloid neoplasms. J Med Genet. 49:422–8. DOI: 10.1136/jmedgenet-2011-100674. PMID: 22652532.
Article15. Voso MT, Fabiani E, Zang Z, Fianchi L, Falconi G, Padella A, et al. 2015; Fanconi anemia gene variants in therapy-related myeloid neoplasms. Blood Cancer J. 5:e323. DOI: 10.1038/bcj.2015.44. PMID: 26140431. PMCID: PMC4526773.
Article16. Yun J, Song H, Kim SM, Kim S, Kwon SR, Lee YE, et al. 2023; Analysis of clinical and genomic profiles of therapy-related myeloid neoplasm in Korea. Hum Genomics. 17:13. DOI: 10.1186/s40246-023-00458-8. PMID: 36814285. PMCID: PMC9948421. PMID: 5a187ce8c60e4562b61c9aecaca68297.
Article17. Greaves M, Maley CC. 2012; Clonal evolution in cancer. Nature. 481:306–13. DOI: 10.1038/nature10762. PMID: 22258609. PMCID: PMC3367003.
Article18. Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. 2017; Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the association for molecular pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 19:4–23. DOI: 10.1016/j.jmoldx.2016.10.002. PMID: 27993330. PMCID: PMC5707196.
Article19. Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. 2000; BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 14:927–39. DOI: 10.1101/gad.14.8.927. PMID: 10783165. PMCID: PMC316544.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Genomic testing for germline predisposition to hematologic malignancies
- Multiple Primary Cancers Including Colorectal Cancer
- Constitutional Mismatch Repair Deficiency, the Most Aggressive Cancer Predisposition Syndrome : Clinical Presentation, Surveillance, and Management
- Multiple Primary Carcinoma Associated with Gynecologic Malignancies
- Extracorporeal Life Support in Patients with Hematologic Malignancies: A Single Center Experience