J Korean Med Sci.
2024 Jul;39(26):e200. 10.3346/jkms.2024.39.e200.
Machine Learning-Based Identification of Diagnostic Biomarkers for Korean Male Sarcopenia Through Integrative DNA Methylation and Methylation Risk Score: From the Korean Genomic Epidemiology Study (KoGES)
- Affiliations
-
- 1Health and Exercise Science Laboratory, Institute of Sport Science, Department of Physical Education, Seoul National University, Seoul, Korea
- 2Institute on Aging, Seoul National University, Seoul, Korea
- KMID: 2557775
- DOI: http://doi.org/10.3346/jkms.2024.39.e200
Abstract
- Background
Sarcopenia, characterized by a progressive decline in muscle mass, strength, and function, is primarily attributable to aging. DNA methylation, influenced by both genetic predispositions and environmental exposures, plays a significant role in sarcopenia occurrence. This study employed machine learning (ML) methods to identify differentially methylated probes (DMPs) capable of diagnosing sarcopenia in middle-aged individuals. We also investigated the relationship between muscle strength, muscle mass, age, and sarcopenia risk as reflected in methylation profiles.
Methods
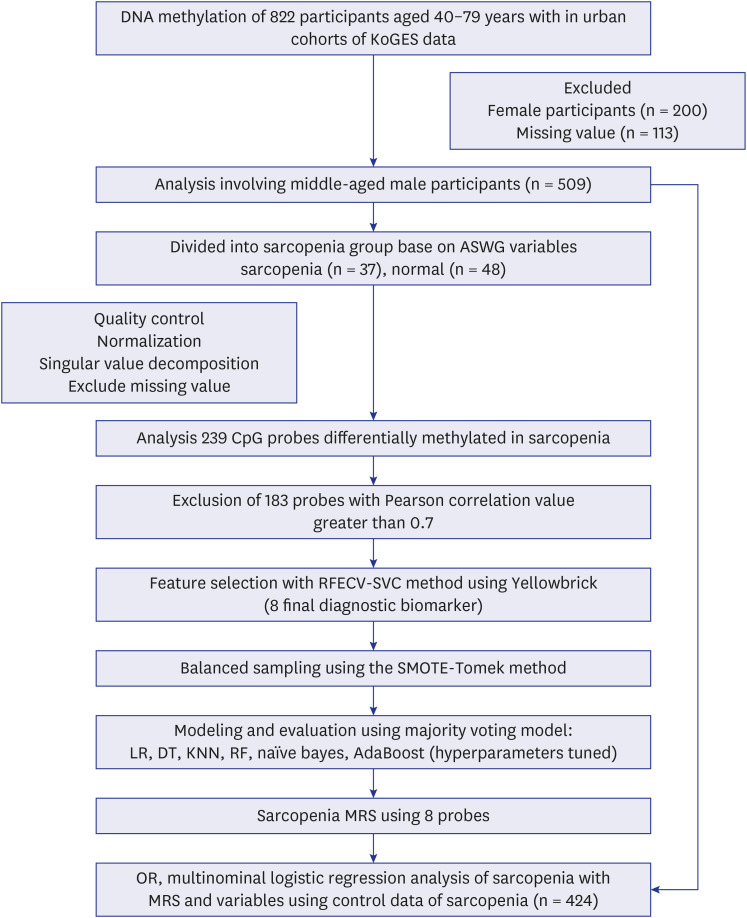
Data from 509 male participants in the urban cohort of the Korean Genome Epidemiology Study_Health Examinee study were categorized into quartile groups based on the sarcopenia criteria for appendicular skeletal muscle index (ASMI) and handgrip strength (HG). To identify diagnostic biomarkers for sarcopenia, we used recursive feature elimination with cross validation (RFECV), to pinpoint DMPs significantly associated with sarcopenia. An ensemble model, leveraging majority voting, was utilized for evaluation. Furthermore, a methylation risk score (MRS) was calculated, and its correlation with muscle strength, function, and age was assessed using likelihood ratio analysis and multinomial logistic regression.
Results
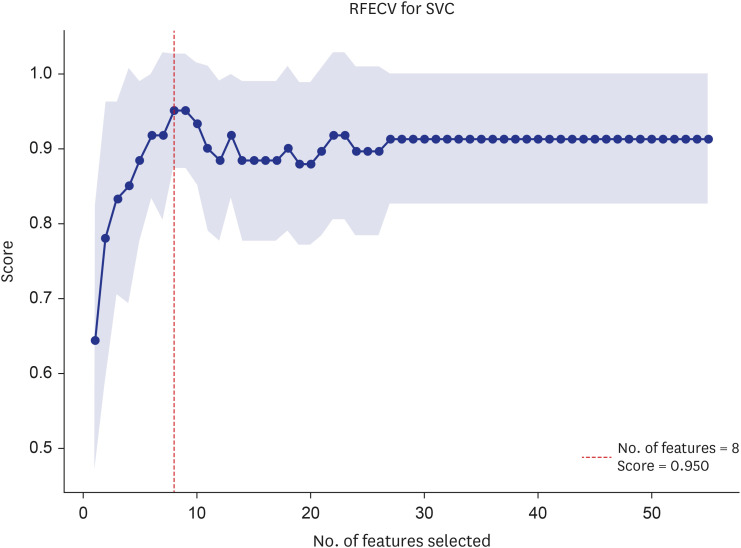
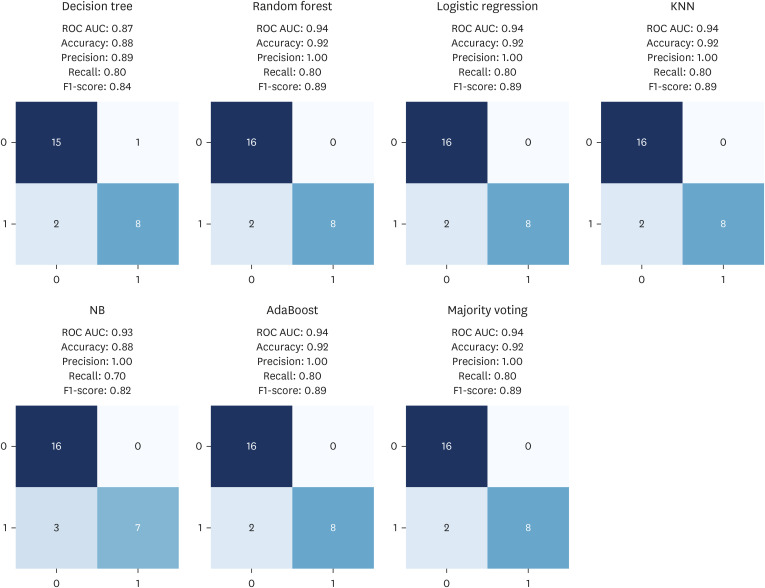
Participants were classified into two groups based on quartile thresholds: sarcopenia (n = 37) with ASMI and HG in the lowest quartile, and normal ranges (n = 48) in the highest. In total, 238 DMPs were identified and eight probes were selected using RFECV. These DMPs were used to build an ensemble model with robust diagnostic capabilities for sarcopenia, as evidenced by an area under the receiver operating characteristic curve of 0.94. Based on eight probes, the MRS was calculated and then validated by analyzing age, HG, and ASMI among the control group (n = 424). Age was positively correlated with high MRS (coefficient, 1.2494; odds ratio [OR], 3.4882), whereas ASMI and HG were negatively correlated with high MRS (ASMI coefficient, −0.4275; OR, 0.6521; HG coefficient, −0.3116; OR, 0.7323).
Conclusion
Overall, this study identified key epigenetic markers of sarcopenia in Korean males and developed a ML model with high diagnostic accuracy for sarcopenia. The MRS also revealed significant correlations between these markers and age, HG, and ASMI. These findings suggest that both diagnostic models and the MRS can play an important role in managing sarcopenia in middle-aged populations.
Keyword
Figure
Reference
-
1. Chen LK, Liu LK, Woo J, Assantachai P, Auyeung TW, Bahyah KS, et al. Sarcopenia in Asia: consensus report of the Asian working group for sarcopenia. J Am Med Dir Assoc. 2014; 15(2):95–101. PMID: 24461239.2. Bijlsma AY, Meskers CG, Ling CH, Narici M, Kurrle SE, Cameron ID, et al. Defining sarcopenia: the impact of different diagnostic criteria on the prevalence of sarcopenia in a large middle aged cohort. Age (Dordr). 2013; 35(3):871–881. PMID: 22314402.3. Ryan AS, Ivey FM, Serra MC, Hartstein J, Hafer-Macko CE. Sarcopenia and physical function in middle-aged and older stroke survivors. Arch Phys Med Rehabil. 2017; 98(3):495–499. PMID: 27530769.4. Du Y, Wang X, Xie H, Zheng S, Wu X, Zhu X, et al. Sex differences in the prevalence and adverse outcomes of sarcopenia and sarcopenic obesity in community dwelling elderly in East China using the AWGS criteria. BMC Endocr Disord. 2019; 19(1):109. PMID: 31653213.5. Chew STH, Tey SL, Yalawar M, Liu Z, Baggs G, How CH, et al. Prevalence and associated factors of sarcopenia in community-dwelling older adults at risk of malnutrition. BMC Geriatr. 2022; 22(1):997. PMID: 36564733.6. Kim M, Won CW. Sarcopenia in Korean community-dwelling adults aged 70 years and older: application of screening and diagnostic tools from the Asian working group for sarcopenia 2019 update. J Am Med Dir Assoc. 2020; 21(6):752–758. PMID: 32386844.7. Haren MT, Banks WA, Perry Iii HM, Patrick P, Malmstrom TK, Miller DK, et al. Predictors of serum testosterone and DHEAS in African-American men. Int J Androl. 2008; 31(1):50–59. PMID: 18190426.8. Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer. 2011; 2(6):607–617. PMID: 21941617.9. Kennedy BK, Berger SL, Brunet A, Campisi J, Cuervo AM, Epel ES, et al. Geroscience: linking aging to chronic disease. Cell. 2014; 159(4):709–713. PMID: 25417146.10. Jordan MI, Mitchell TM. Machine learning: trends, perspectives, and prospects. Science. 2015; 349(6245):255–260. PMID: 26185243.11. Burns JE, Yao J, Chalhoub D, Chen JJ, Summers RM. A machine learning algorithm to estimate sarcopenia on abdominal CT. Acad Radiol. 2020; 27(3):311–320. PMID: 31126808.12. Kang YJ, Yoo JI, Ha YC. Sarcopenia feature selection and risk prediction using machine learning: a cross-sectional study. Medicine (Baltimore). 2019; 98(43):e17699. PMID: 31651901.13. Ao C, Gao L, Yu L. Research progress in predicting DNA methylation modifications and the relation with human diseases. Curr Med Chem. 2022; 29(5):822–836. PMID: 34533438.14. Kim Y, Han BG. KoGES group. Cohort profile: the Korean Genome and Epidemiology Study (KoGES) consortium. Int J Epidemiol. 2017; 46(2):e20. PMID: 27085081.15. Chen LK, Woo J, Assantachai P, Auyeung TW, Chou MY, Iijima K, et al. Asian Working Group for Sarcopenia: 2019 consensus update on sarcopenia diagnosis and treatment. J Am Med Dir Assoc. 2020; 21(3):300–307.e2. PMID: 32033882.16. Wen X, Wang M, Jiang CM, Zhang YM. Anthropometric equation for estimation of appendicular skeletal muscle mass in Chinese adults. Asia Pac J Clin Nutr. 2011; 20(4):551–556. PMID: 22094840.17. Wigodski S, Carrasco F, Bunout D, Barrera G, Hirsch S, de la Maza MP. Sarcopenia: the need to establish different cutoff points of fat-free mass for the Chilean population. Nutrition. 2019; 57:217–224. PMID: 30184515.18. Kim JW, Yoon JS, Kim EJ, Hong HL, Kwon HH, Jung CY, et al. Prognostic implication of baseline sarcopenia for length of hospital stay and survival in patients with coronavirus disease 2019. J Gerontol A Biol Sci Med Sci. 2021; 76(8):e110–e116. PMID: 33780535.19. Linge J, Petersson M, Forsgren MF, Sanyal AJ, Dahlqvist Leinhard O. Adverse muscle composition predicts all-cause mortality in the UK Biobank imaging study. J Cachexia Sarcopenia Muscle. 2021; 12(6):1513–1526. PMID: 34713982.20. Bolte FJ, McTavish S, Wakefield N, Shantzer L, Hubbard C, Krishnaraj A, et al. Association of sarcopenia with survival in advanced NSCLC patients receiving concurrent immunotherapy and chemotherapy. Front Oncol. 2022; 12:986236. PMID: 36212442.21. Oshita K, Myotsuzono R, Tashiro T. Association between normal weight obesity and skeletal muscle mass index in female university students with past exercise habituation. J Funct Morphol Kinesiol. 2022; 7(4):92. PMID: 36278753.22. Tian Y, Morris TJ, Webster AP, Yang Z, Beck S, Feber A, et al. ChAMP: updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics. 2017; 33(24):3982–3984. PMID: 28961746.23. Unruh D, Zewde M, Buss A, Drumm MR, Tran AN, Scholtens DM, et al. Methylation and transcription patterns are distinct in IDH mutant gliomas compared to other IDH mutant cancers. Sci Rep. 2019; 9(1):8946. PMID: 31222125.24. Chandra A, Senapati S, Roy S, Chatterjee G, Chatterjee R. Epigenome-wide DNA methylation regulates cardinal pathological features of psoriasis. Clin Epigenetics. 2018; 10(1):108. PMID: 30092825.25. Ringh MV, Hagemann-Jensen M, Needhamsen M, Kular L, Breeze CE, Sjöholm LK, et al. Tobacco smoking induces changes in true DNA methylation, hydroxymethylation and gene expression in bronchoalveolar lavage cells. EBioMedicine. 2019; 46:290–304. PMID: 31303497.26. Huang X, Zhang L, Wang B, Li F, Zhang Z. Feature clustering based support vector machine recursive feature elimination for gene selection. Appl Intell. 2018; 48(3):594–607.27. Bengfort B, Bilbro R. Yellowbrick: visualizing the scikit-learn model selection process. J Open Source Softw. 2019; 4(35):1075.28. Batista GEAPA, Bazzan ALC, Monard MC. Balancing training data for automated annotation of keywords: a case study. Wob. 2003; 3:10–18.29. Thompson M, Hill BL, Rakocz N, Chiang JN, Geschwind D, Sankararaman S, et al. Methylation risk scores are associated with a collection of phenotypes within electronic health record systems. NPJ Genom Med. 2022; 7(1):50. PMID: 36008412.30. Day K, Waite LL, Thalacker-Mercer A, West A, Bamman MM, Brooks JD, et al. Differential DNA methylation with age displays both common and dynamic features across human tissues that are influenced by CpG landscape. Genome Biol. 2013; 14(9):R102. PMID: 24034465.31. Turner DC, Gorski PP, Maasar MF, Seaborne RA, Baumert P, Brown AD, et al. DNA methylation across the genome in aged human skeletal muscle tissue and muscle-derived cells: the role of HOX genes and physical activity. Sci Rep. 2020; 10(1):15360. PMID: 32958812.32. Zykovich A, Hubbard A, Flynn JM, Tarnopolsky M, Fraga MF, Kerksick C, et al. Genome-wide DNA methylation changes with age in disease-free human skeletal muscle. Aging Cell. 2014; 13(2):360–366. PMID: 24304487.33. Marx SO, Marks AR. Dysfunctional ryanodine receptors in the heart: new insights into complex cardiovascular diseases. J Mol Cell Cardiol. 2013; 58:225–231. PMID: 23507255.34. Marks AR. Targeting ryanodine receptors to treat human diseases. J Clin Invest. 2023; 133(2):e162891. PMID: 36647824.35. Frattini A, Pangrazio A, Susani L, Sobacchi C, Mirolo M, Abinun M, et al. Chloride channel ClCN7 mutations are responsible for severe recessive, dominant, and intermediate osteopetrosis. J Bone Miner Res. 2003; 18(10):1740–1747. PMID: 14584882.36. Rössler U, Hennig AF, Stelzer N, Bose S, Kopp J, Søe K, et al. Efficient generation of osteoclasts from human induced pluripotent stem cells and functional investigations of lethal CLCN7-related osteopetrosis. J Bone Miner Res. 2021; 36(8):1621–1635. PMID: 33905594.37. Wang X, Wang Y, Xu T, Fan Y, Ding Y, Qian J. A novel compound heterozygous mutation of the CLCN7 gene is associated with autosomal recessive osteopetrosis. Front Pediatr. 2023; 11:978879. PMID: 37168803.38. El-Gazzar A, Voraberger B, Rauch F, Mairhofer M, Schmidt K, Guillemyn B, et al. Bi-allelic mutation in SEC16B alters collagen trafficking and increases ER stress. EMBO Mol Med. 2023; 15(4):e16834. PMID: 36916446.39. Bennett JL, Pratt AG, Dodds R, Sayer AA, Isaacs JD. Rheumatoid sarcopenia: loss of skeletal muscle strength and mass in rheumatoid arthritis. Nat Rev Rheumatol. 2023; 19(4):239–251. PMID: 36801919.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- DNA Methylation in Development
- DNA methylation: a cause and consequence of type 2 diabetes
- A Two-DNA Methylation Signature to Improve Prognosis Prediction of Clear Cell Renal Cell Carcinoma
- A Visualization Tool for Computational Analysis of DNA Methylation Level Using Bisulfite Sequencing Data
- DNA Methylation Profiles of Blood Cells Are Distinct between Early-Onset Obese and Control Individuals