Korean J Physiol Pharmacol.
2024 Jul;28(4):361-377. 10.4196/kjpp.2024.28.4.361.
Analysis of the mechanism of fibrauretine alleviating Alzheimer's disease based on transcriptomics and proteomics
- Affiliations
-
- 1College of Chinese Medicinal Materials, Jilin Agricultural University, Changchun 130118, China
- 2Key Laboratory of Animal Production, Product Quality and Security, Ministry of Education of China, Changchun 130118, China
- 3Jilin Provincial Engineering Research Center for Efficient Breeding and Product Development of Sika Deer of China, Changchun 130118, China
- KMID: 2557208
- DOI: http://doi.org/10.4196/kjpp.2024.28.4.361
Abstract
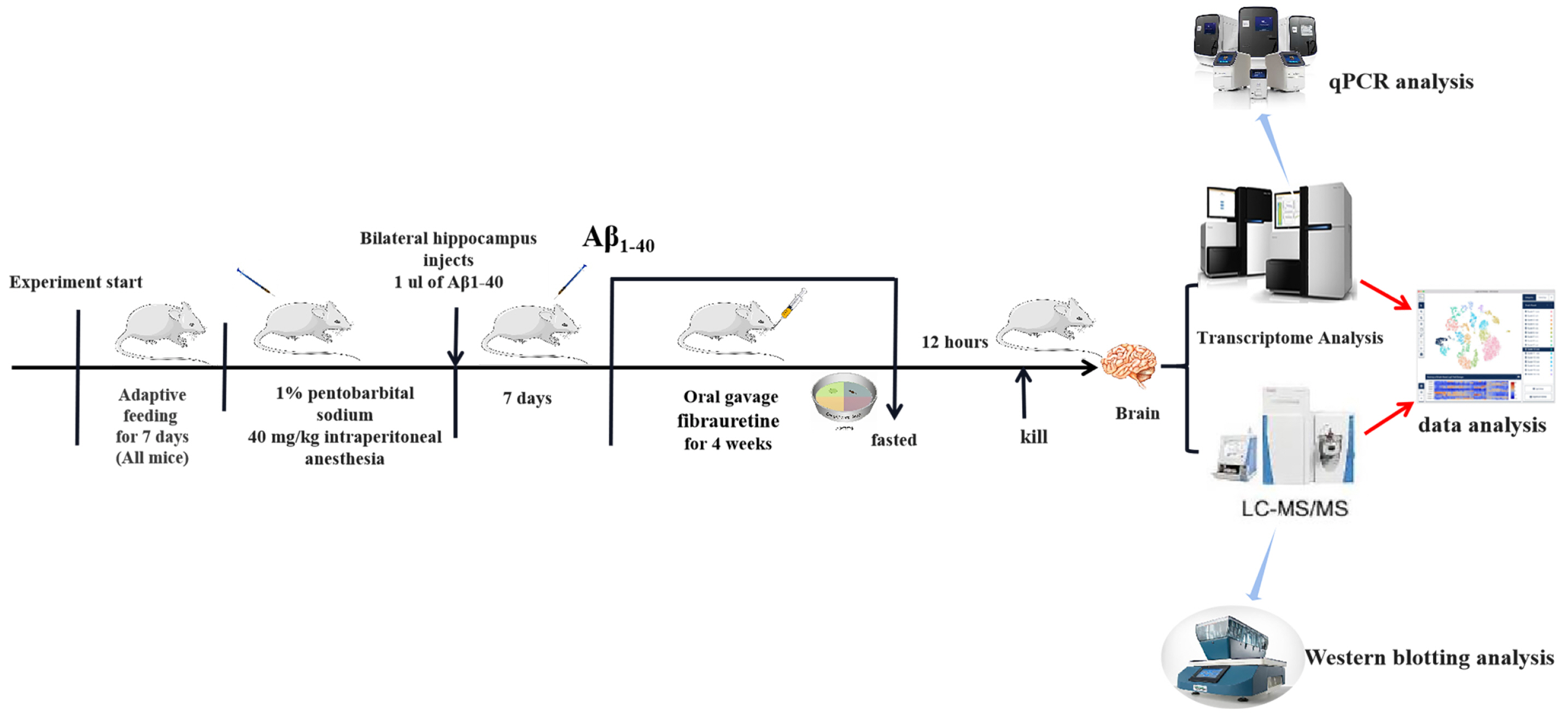
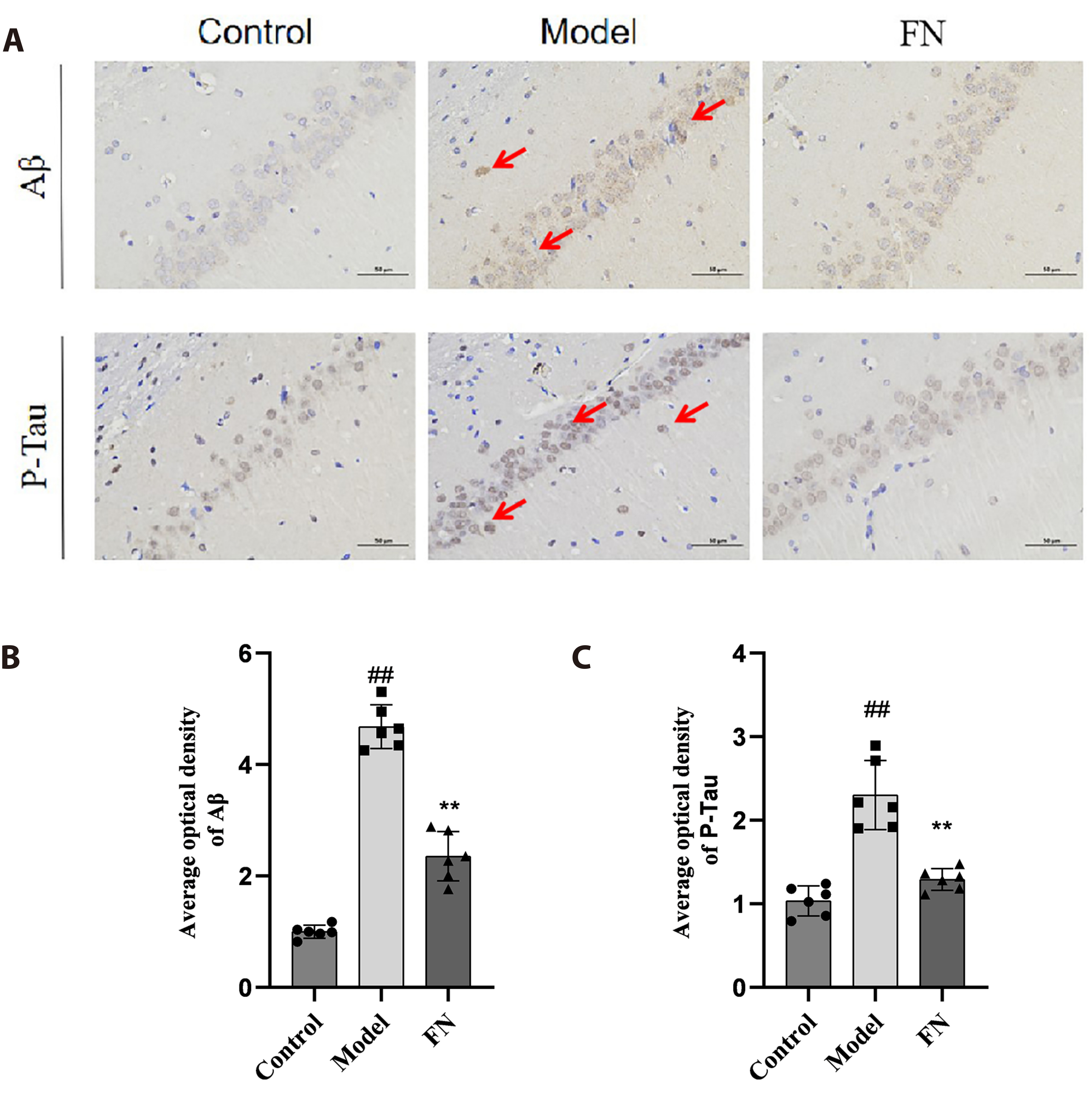
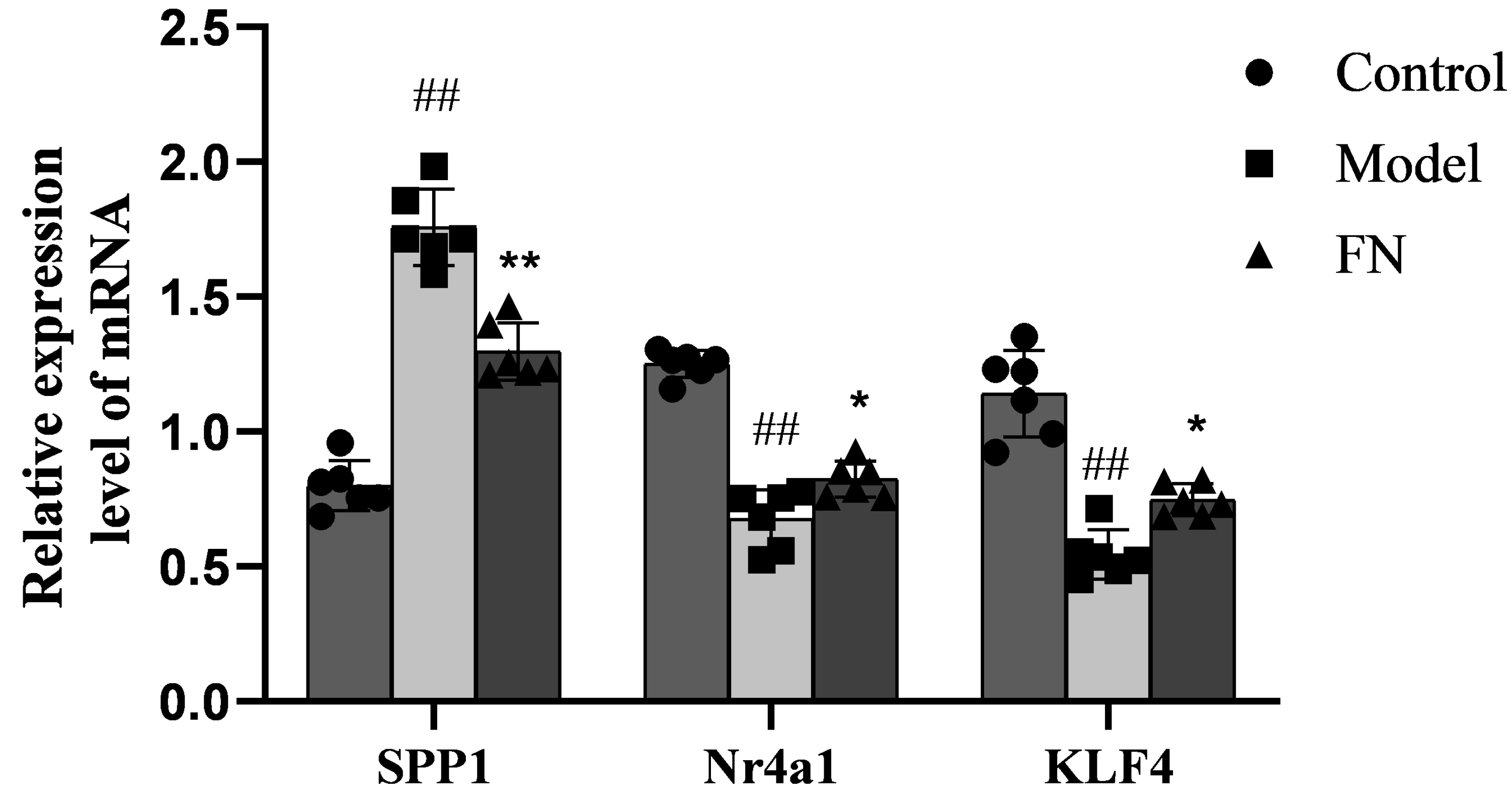
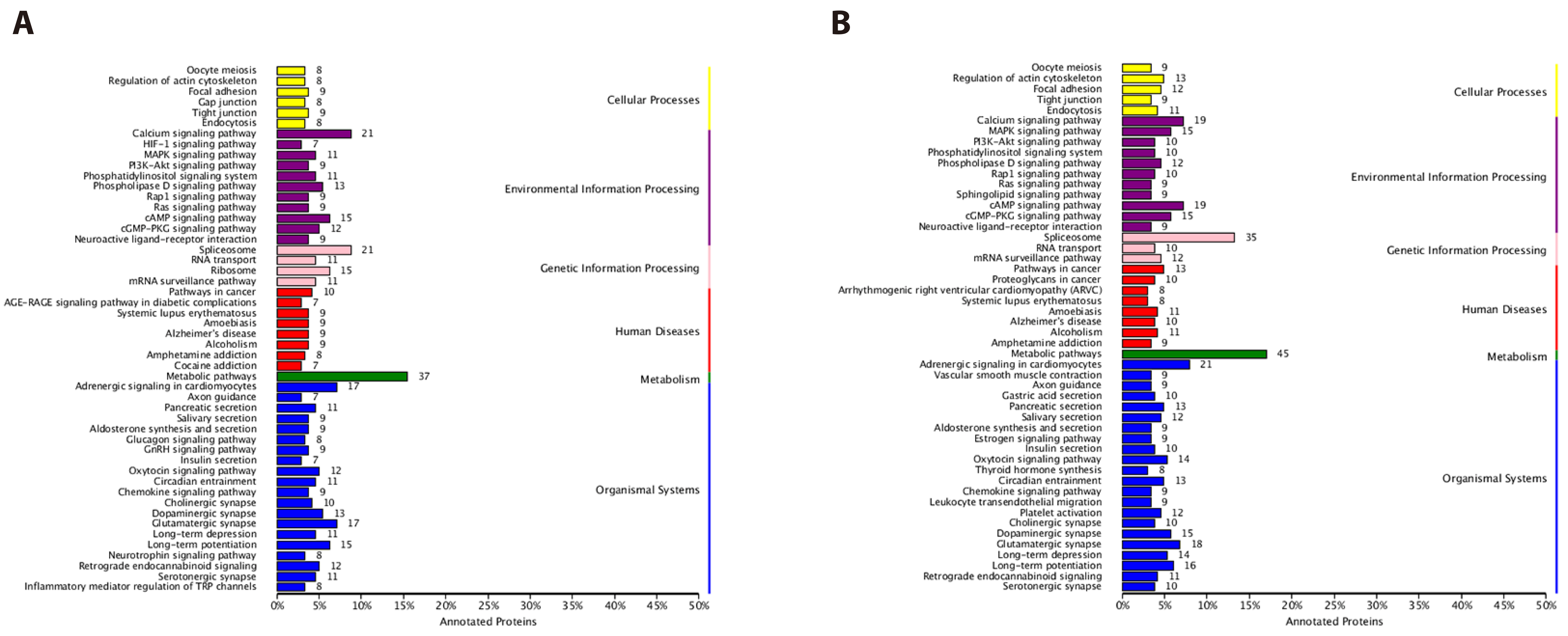
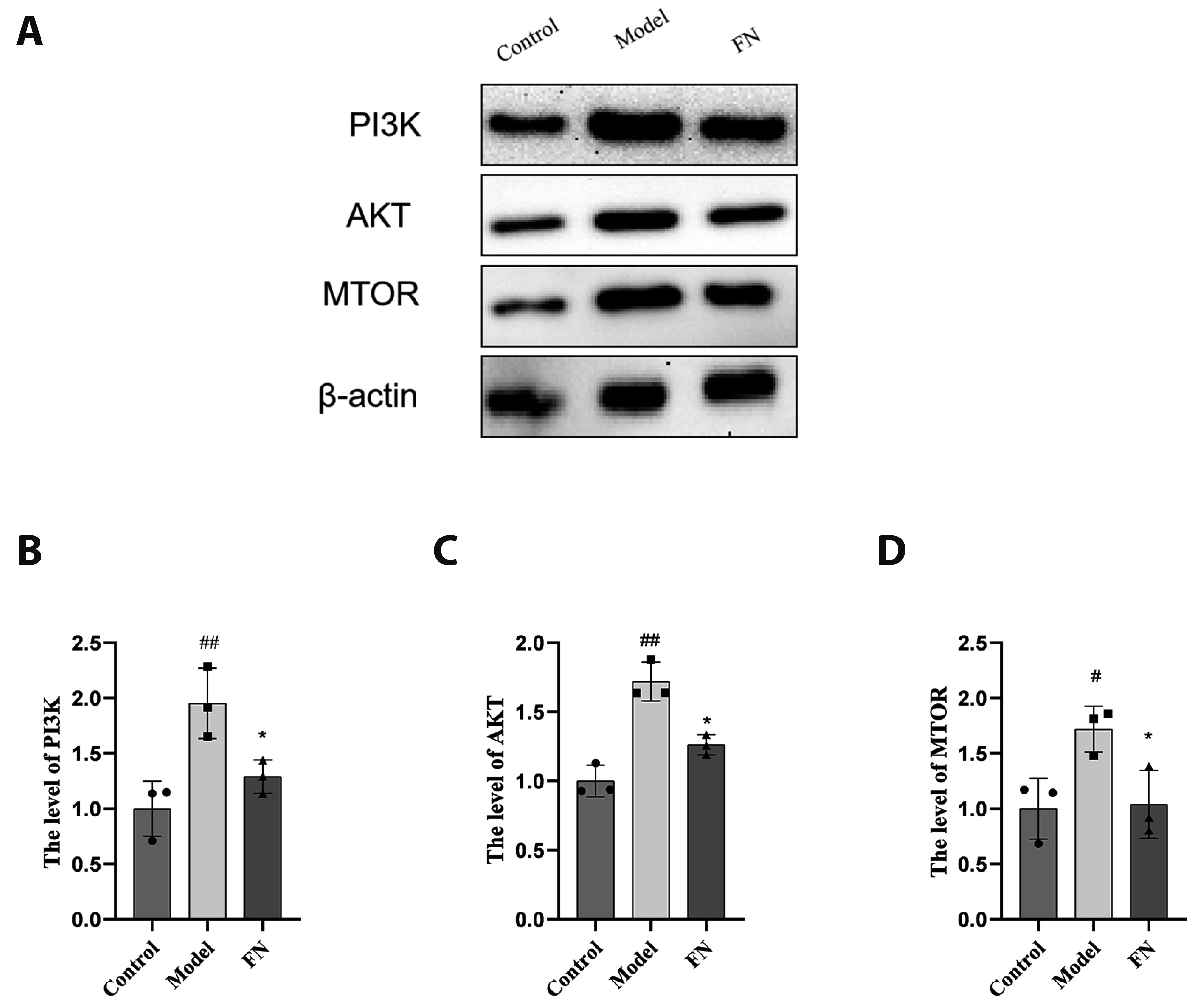
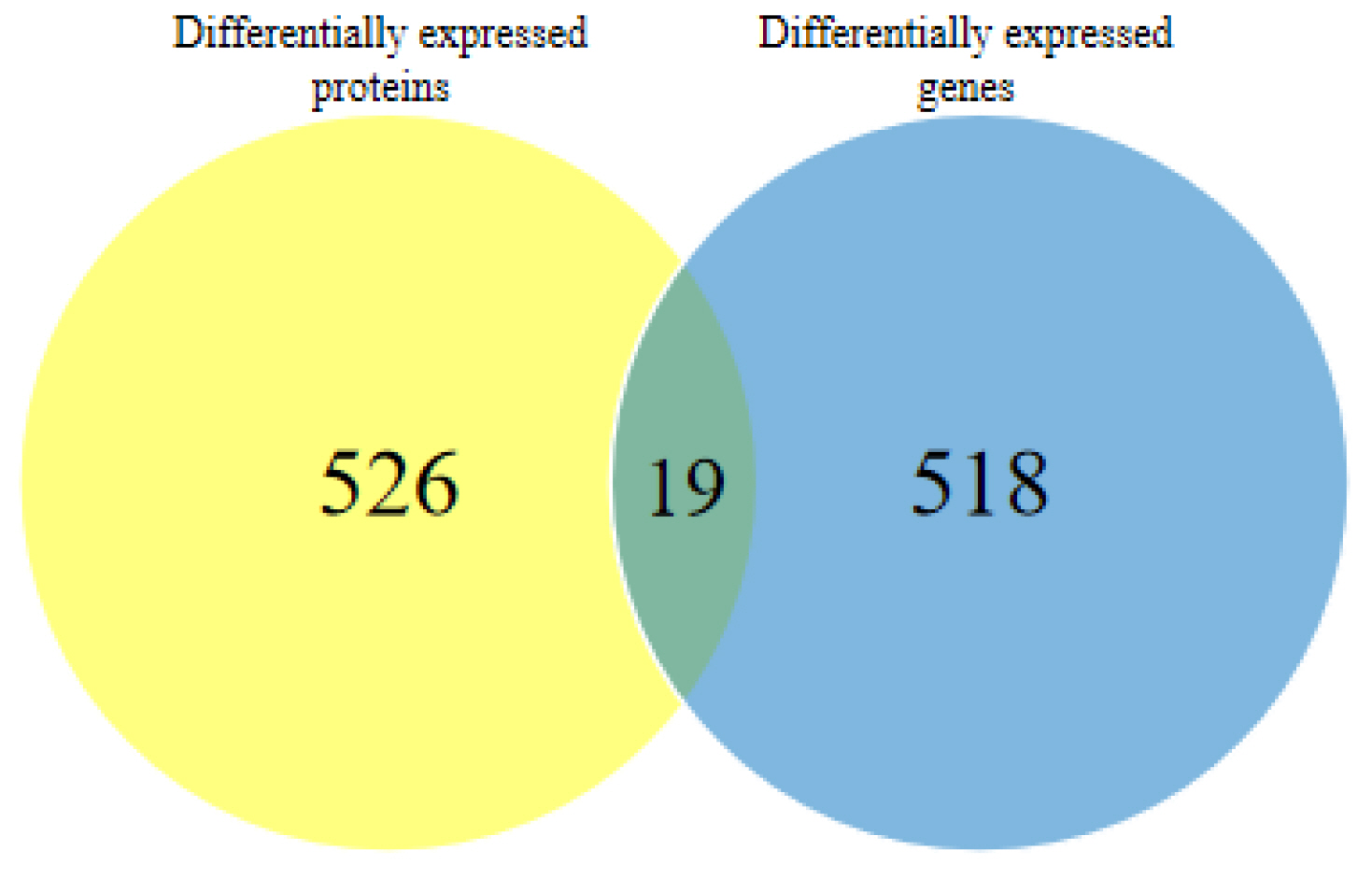
- The dried rattan stem of the Fibraurea Recisa Pierre plant contains the active ingredient known as fibrauretine (FN). Although it greatly affects Alzheimer's disease (AD), the mechanism of their effects still remains unclear. Proteomics and transcriptomics analysis methods were used in this study to determine the mechanism of FN in the treatment of AD. AD model is used through bilateral hippocampal injection of Aβ1-40. After successful modeling, FN was given for 30 days. The results showed that FN could improve the cognitive dysfunction of AD model rats, reduce the expression of Aβ and P-Tau, increase the content of acetylcholine and reduce the activity of acetylcholinesterase. The Kyoto Encyclopedia of Genes and Genomes enriched differentially expressed genes and proteins are involved in signaling pathways including metabolic pathway, AD, pathway in cancer, PI3K-AKT signaling pathway, and cAMP signaling pathway. Transcriptomics and proteomics sequencing resulted in 19 differentially expressed genes and proteins. Finally, in contrast to the model group, after FN treatment, the protein expressions and genes associated with the PI3K-AKT pathway were significantly improved in RT-qPCR and Western blot and assays. This is consistent with the findings of transcriptomic and proteomic analyses. Our study found that, FN may improve some symptoms of AD model rats through PI3K-AKT signaling pathway.
Keyword
Figure
Reference
-
1. Scheltens P, De Strooper B, Kivipelto M, Holstege H, Chételat G, Teunissen CE, Cummings J, van der Flier WM. 2021; Alzheimer's disease. Lancet. 397:1577–1590. DOI: 10.1016/S0140-6736(20)32205-4. PMID: 33667416.2. Dujardin P, Vandenbroucke RE, Van Hoecke L. 2022; Fighting fire with fire: the immune system might be key in our fight against Alzheimer's disease. Drug Discov Today. 27:1261–1283. DOI: 10.1016/j.drudis.2022.01.004. PMID: 35032668.3. Ye JY, Li L, Hao QM, Qin Y, Ma CS. 2020; β-Sitosterol treatment attenuates cognitive deficits and prevents amyloid plaque deposition in amyloid protein precursor/presenilin 1 mice. Korean J Physiol Pharmacol. 24:39–46. DOI: 10.4196/kjpp.2020.24.1.39. PMID: 31908573. PMCID: PMC6940499.4. Xing Z, He Z, Wang S, Yan Y, Zhu H, Gao Y, Zhao Y, Zhang L. 2018; Ameliorative effects and possible molecular mechanisms of action of fibrauretine from Fibraurea recisa Pierre on d-galactose/AlCl3-mediated Alzheimer's disease. RSC Adv. 8:31646–31657. DOI: 10.1039/C8RA05356A. PMID: 35548215. PMCID: PMC9085853.5. Wang Y, Lin Y, Wang L, Zhan H, Luo X, Zeng Y, Wu W, Zhang X, Wang F. 2020; TREM2 ameliorates neuroinflammatory response and cognitive impairment via PI3K/AKT/FoxO3a signaling pathway in Alzheimer's disease mice. Aging (Albany NY). 12:20862–20879. DOI: 10.18632/aging.104104. PMID: 33065553. PMCID: PMC7655179.6. Sebastian Monasor L, Müller SA, Colombo AV, Tanrioever G, König J, Roth S, Liesz A, Berghofer A, Piechotta A, Prestel M, Saito T, Saido TC, Herms J, Willem M, Haass C, Lichtenthaler SF, Tahirovic S. 2020; Fibrillar Aβ triggers microglial proteome alterations and dysfunction in Alzheimer mouse models. Elife. 9:e54083. DOI: 10.7554/eLife.54083. PMID: 32510331. PMCID: PMC7279888.7. Sur B, Lee B. 2022; Myricetin prevents sleep deprivation-induced cognitive impairment and neuroinflammation in rat brain via regulation of brain-derived neurotropic factor. Korean J Physiol Pharmacol. 26:415–425. DOI: 10.4196/kjpp.2022.26.6.415. PMID: 36302617. PMCID: PMC9614391.8. Celik Topkara K, Kilinc E, Cetinkaya A, Saylan A, Demir S. 2022; Therapeutic effects of carvacrol on beta-amyloid-induced impairments in in vitro and in vivo models of Alzheimer's disease. Eur J Neurosci. 56:5714–5726. DOI: 10.1111/ejn.15565. PMID: 34904309.9. Lee B, Yeom M, Shim I, Lee H, Hahm DH. 2020; Inhibitory effect of carvacrol on lipopolysaccharide-induced memory impairment in rats. Korean J Physiol Pharmacol. 24:27–37. DOI: 10.4196/kjpp.2020.24.1.27. PMID: 31908572. PMCID: PMC6940503.10. Kirino T. 1982; Delayed neuronal death in the gerbil hippocampus following ischemia. Brain Res. 239:57–69. DOI: 10.1016/0006-8993(82)90833-2. PMID: 7093691.11. Shukla PK, Meena AS, Manda B, Gomes-Solecki M, Dietrich P, Dragatsis I, Rao R. 2018; Lactobacillus plantarum prevents and mitigates alcohol-induced disruption of colonic epithelial tight junctions, endotoxemia, and liver damage by an EGF receptor-dependent mechanism. FASEB J. 32:fj201800351R. DOI: 10.1096/fj.201800351R. PMID: 29912589. PMCID: PMC6181630.12. Yang X, Guan Y, Yan B, Xie Y, Zhou M, Wu Y, Yao L, Qiu X, Yan F, Chen Y, Huang L. 2021; Evidence-based complementary and alternative medicine bioinformatics approach through network pharmacology and molecular docking to determine the molecular mechanisms of Erjing pill in Alzheimer's disease. Exp Ther Med. 22:1252. DOI: 10.3892/etm.2021.10687. PMID: 34539848. PMCID: PMC8438686.13. Park GW, Hwang H, Kim KH, Lee JY, Lee HK, Park JY, Ji ES, Park SR, Yates JR 3rd, Kwon KH, Park YM, Lee HJ, Paik YK, Kim JY, Yoo JS. 2016; Integrated proteomic pipeline using multiple search engines for a proteogenomic study with a controlled protein false discovery rate. J Proteome Res. 15:4082–4090. DOI: 10.1021/acs.jproteome.6b00376. PMID: 27537616.14. Bloom GS. 2014; Amyloid-β and tau: the trigger and bullet in Alzheimer disease pathogenesis. JAMA Neurol. 71:505–508. DOI: 10.1001/jamaneurol.2013.5847. PMID: 24493463.15. Furukawa A, Kawamoto Y, Chiba Y, Takei S, Hasegawa-Ishii S, Kawamura N, Yoshikawa K, Hosokawa M, Oikawa S, Kato M, Shimada A. 2011; Proteomic identification of hippocampal proteins vulnerable to oxidative stress in excitotoxin-induced acute neuronal injury. Neurobiol Dis. 43:706–714. DOI: 10.1016/j.nbd.2011.05.024. PMID: 21669285.16. Pedrero-Prieto CM, García-Carpintero S, Frontiñán-Rubio J, Llanos-González E, Aguilera García C, Alcaín FJ, Lindberg I, Durán-Prado M, Peinado JR, Rabanal-Ruiz Y. 2020; A comprehensive systematic review of CSF proteins and peptides that define Alzheimer's disease. Clin Proteomics. 17:21. DOI: 10.1186/s12014-020-09276-9. PMID: 32518535. PMCID: PMC7273668.17. Tsuboyama M, Iqbal MA. 2021; CHL1 deletion is associated with cognitive and language disabilities - Case report and review of literature. Mol Genet Genomic Med. 9:e1725. DOI: 10.1002/mgg3.1725. PMID: 34056867. PMCID: PMC8372067.18. Zhuang X, Zhang G, Bao M, Jiang G, Wang H, Li S, Wang Z, Sun X. 2023; Development of a novel immune infiltration-related diagnostic model for Alzheimer's disease using bioinformatic strategies. Front Immunol. 14:1147501. DOI: 10.3389/fimmu.2023.1147501. PMID: 37545529. PMCID: PMC10400274.19. Maes M, DeVos N, Wauters A, Demedts P, Maurits VW, Neels H, Bosmans E, Altamura C, Lin A, Song C, Vandenbroucke M, Scharpe S. 1999; Inflammatory markers in younger vs elderly normal volunteers and in patients with Alzheimer's disease. J Psychiatr Res. 33:397–405. DOI: 10.1016/S0022-3956(99)00016-3. PMID: 10504008.20. Bai Y, Li L, Dong B, Ma W, Chen H, Yu Y. 2022; Phosphorylation-mediated PI3K-Art signalling pathway as a therapeutic mechanism in the hydrogen-induced alleviation of brain injury in septic mice. J Cell Mol Med. 26:5713–5727. DOI: 10.1111/jcmm.17568. PMID: 36308410. PMCID: PMC9667523.21. Lissanu Deribe Y. 2016; Interplay between PREX2 mutations and the PI3K pathway and its effect on epigenetic regulation of gene expression in NRAS-mutant melanoma. Small GTPases. 7:178–185. DOI: 10.1080/21541248.2016.1178366. PMID: 27111337. PMCID: PMC5003543.22. Zhang Q, Li L, Lai Y, Zhao T. 2020; Silencing of SPP1 suppresses progression of tongue cancer by mediating the PI3K/Akt signaling pathway. Technol Cancer Res Treat. 19:1533033820971306. DOI: 10.1177/1533033820971306. PMID: 33174521. PMCID: PMC7672768.23. Zhang Y, Li S, Cui X, Wang Y. 2023; microRNA-944 inhibits breast cancer cell proliferation and promotes cell apoptosis by reducing SPP1 through inactivating the PI3K/Akt pathway. Apoptosis. 28:1546–1563. DOI: 10.1007/s10495-023-01870-0. PMID: 37486406.24. Briggs R, Kennelly SP, O'Neill D. 2016; Drug treatments in Alzheimer's disease. Clin Med (Lond). 16:247–253. DOI: 10.7861/clinmedicine.16-3-247. PMID: 27251914. PMCID: PMC5922703.25. Jia W, Su Q, Cheng Q, Peng Q, Qiao A, Luo X, Zhang J, Wang Y. 2021; Neuroprotective effects of palmatine via the enhancement of antioxidant defense and small heat shock protein expression in Aβ-transgenic Caenorhabditis elegans. Oxid Med Cell Longev. 2021:9966223. DOI: 10.1155/2021/9966223. PMID: 34567416. PMCID: PMC8460366.26. Pei H, Zeng J, He Z, Zong Y, Zhao Y, Li J, Chen W, Du R. 2023; Palmatine ameliorates LPS-induced HT-22 cells and mouse models of depression by regulating apoptosis and oxidative stress. J Biochem Mol Toxicol. 37:e23225. DOI: 10.1002/jbt.23225. PMID: 36169195.27. Millan MJ. 2017; Linking deregulation of non-coding RNA to the core pathophysiology of Alzheimer's disease: an integrative review. Prog Neurobiol. 156:1–68. DOI: 10.1016/j.pneurobio.2017.03.004. PMID: 28322921.28. Verheijen J, Sleegers K. 2018; Understanding Alzheimer disease at the interface between genetics and transcriptomics. Trends Genet. 34:434–447. DOI: 10.1016/j.tig.2018.02.007. PMID: 29573818.29. Zhang T, Shen Y, Guo Y, Yao J. 2021; Identification of key transcriptome biomarkers based on a vital gene module associated with pathological changes in Alzheimer's disease. Aging (Albany NY). 13:14940–14967. DOI: 10.18632/aging.203017. PMID: 34031265. PMCID: PMC8221319.30. De Schepper S, Ge JZ, Crowley G, Ferreira LSS, Garceau D, Toomey CE, Sokolova D, Rueda-Carrasco J, Shin SH, Kim JS, Childs T, Lashley T, Burden JJ, Sasner M, Sala Frigerio C, Jung S, Hong S. 2023; Perivascular cells induce microglial phagocytic states and synaptic engulfment via SPP1 in mouse models of Alzheimer's disease. Nat Neurosci. 26:406–415. DOI: 10.1038/s41593-023-01257-z. PMID: 36747024. PMCID: PMC9991912.31. Zhang Y, Zhu H, Liu L, Ma H, Shi Q, Li D, Ju X. 2023; Three-dimensional culture of rat ovarian granulosa cells shows increased SPP1 and FGF7 expression through the PI3K/Akt pathway. Cell Biol Int. 47:1004–1016. DOI: 10.1002/cbin.11998. PMID: 36701359.32. Zhang Z, Yu J. 2018; NR4A1 promotes cerebral ischemia reperfusion injury by repressing Mfn2-mediated mitophagy and inactivating the MAPK-ERK-CREB signaling pathway. Neurochem Res. 43:1963–1977. DOI: 10.1007/s11064-018-2618-4. PMID: 30136162.33. McMorrow JP, Murphy EP. 2011; Inflammation: a role for NR4A orphan nuclear receptors? Biochem Soc Trans. 39:688–693. DOI: 10.1042/BST0390688. PMID: 21428963.34. Paillasse MR, de Medina P. 2015; The NR4A nuclear receptors as potential targets for anti-aging interventions. Med Hypotheses. 84:135–140. DOI: 10.1016/j.mehy.2014.12.003. PMID: 25543265.35. Blacher E, Tsai C, Litichevskiy L, Shipony Z, Iweka CA, Schneider KM, Chuluun B, Heller HC, Menon V, Thaiss CA, Andreasson KI. 2022; Aging disrupts circadian gene regulation and function in macrophages. Nat Immunol. 23:229–236. DOI: 10.1038/s41590-021-01083-0. PMID: 34949832. PMCID: PMC9704320.36. Ji CY, Yuan MX, Chen LJ, Gao HQ, Zhang T, Yin Q. 2022; miR92a represses the viability and migration of nerve cells in Hirschsprung's disease by regulating the KLF4/PI3K/AKT pathway. Acta Neurobiol Exp (Wars). 82:336–346. DOI: 10.55782/ane-2022-032. PMID: 36214716.37. Andreev VP, Petyuk VA, Brewer HM, Karpievitch YV, Xie F, Clarke J, Camp D, Smith RD, Lieberman AP, Albin RL, Nawaz Z, El Hokayem J, Myers AJ. 2012; Label-free quantitative LC-MS proteomics of Alzheimer's disease and normally aged human brains. J Proteome Res. 11:3053–3067. DOI: 10.1021/pr3001546. PMID: 22559202. PMCID: PMC3445701.38. Hui W, Feng Y, Ruihua X, Jiongjie H, Dongan C, Yan S, Shidong Z. 2020; Comparative proteomics analysis indicates that palmatine contributes to transepithelial migration by regulating cellular adhesion. Pharm Biol. 58:646–654. Erratum in: Pharm Biol. 2021;59:546. DOI: 10.1080/13880209.2020.1784961. PMID: 32658562. PMCID: PMC7470081.39. Johnson ECB, Dammer EB, Duong DM, Ping L, Zhou M, Yin L, Higginbotham LA, Guajardo A, White B, Troncoso JC, Thambisetty M, Montine TJ, Lee EB, Trojanowski JQ, Beach TG, Reiman EM, Haroutunian V, Wang M, Schadt E, Zhang B, et al. 2020; Large-scale proteomic analysis of Alzheimer's disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat Med. 26:769–780. DOI: 10.1038/s41591-020-0815-6. PMID: 32284590. PMCID: PMC7405761.40. Zhang S, Sun P, Xiao X, Hu Y, Qian Y, Zhang Q. 2022; MicroRNA-21 promotes epithelial-mesenchymal transition and migration of human bronchial epithelial cells by targeting poly (ADP-ribose) polymerase-1 and activating PI3K/AKT signaling. Korean J Physiol Pharmacol. 26:239–253. DOI: 10.4196/kjpp.2022.26.4.239. PMID: 35766002. PMCID: PMC9247709.41. Long HZ, Cheng Y, Zhou ZW, Luo HY, Wen DD, Gao LC. 2021; PI3K/AKT signal pathway: a target of natural products in the prevention and treatment of Alzheimer's disease and Parkinson's disease. Front Pharmacol. 12:648636. DOI: 10.3389/fphar.2021.648636. PMID: 33935751. PMCID: PMC8082498.