Ewha Med J.
2024 Apr;47(2):e23. 10.12771/emj.2024.e23.
An accurate pediatric bone age prediction model using deep learning and contrast conversion
- Affiliations
-
- 1Department of Medicine, Yonsei University College of Medicine, Seoul, Korea
- 2Medical Physics and Biomedical Engineering Lab (MPBEL), Yonsei University College of Medicine, Seoul, Korea
- 3Department of Radiation Oncology, Heavy Ion Therapy Research Institute, Yonsei Cancer Center, Yonsei University College of Medicine, Seoul, Korea
- 4Ewha Medical Research Institute, Ewha Womans University College of Medicine , Seoul, Korea
- 5Ewha Medical Artificial Intelligence Research Institute, Ewha Womans University College of Medicine, Seoul, Korea
- 6Department of Biomedical Engineering, Ewha Womans University College of Medicine, Seoul, Korea
- KMID: 2556312
- DOI: http://doi.org/10.12771/emj.2024.e23
Abstract
Objectives
This study aimed to develop an accurate pediatric bone age prediction model by utilizing deep learning models and contrast conversion techniques, in order to improve growth assessment and clinical decision-making in clinical practice.
Methods
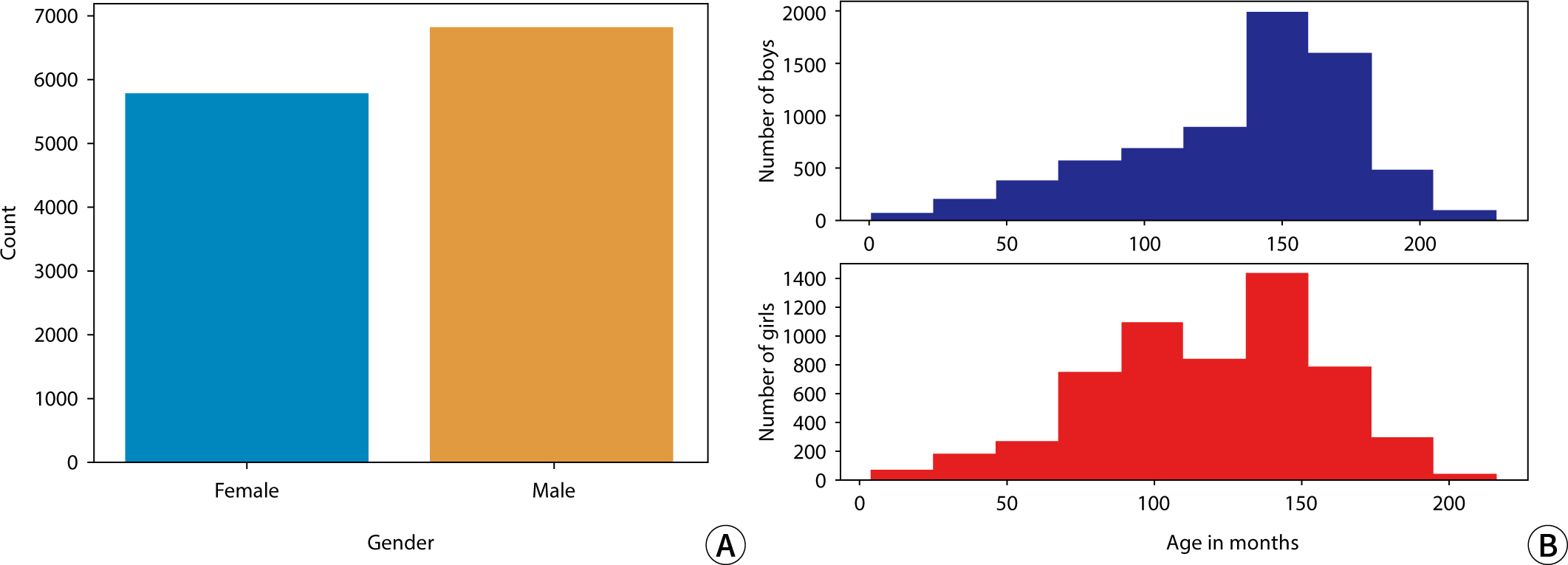
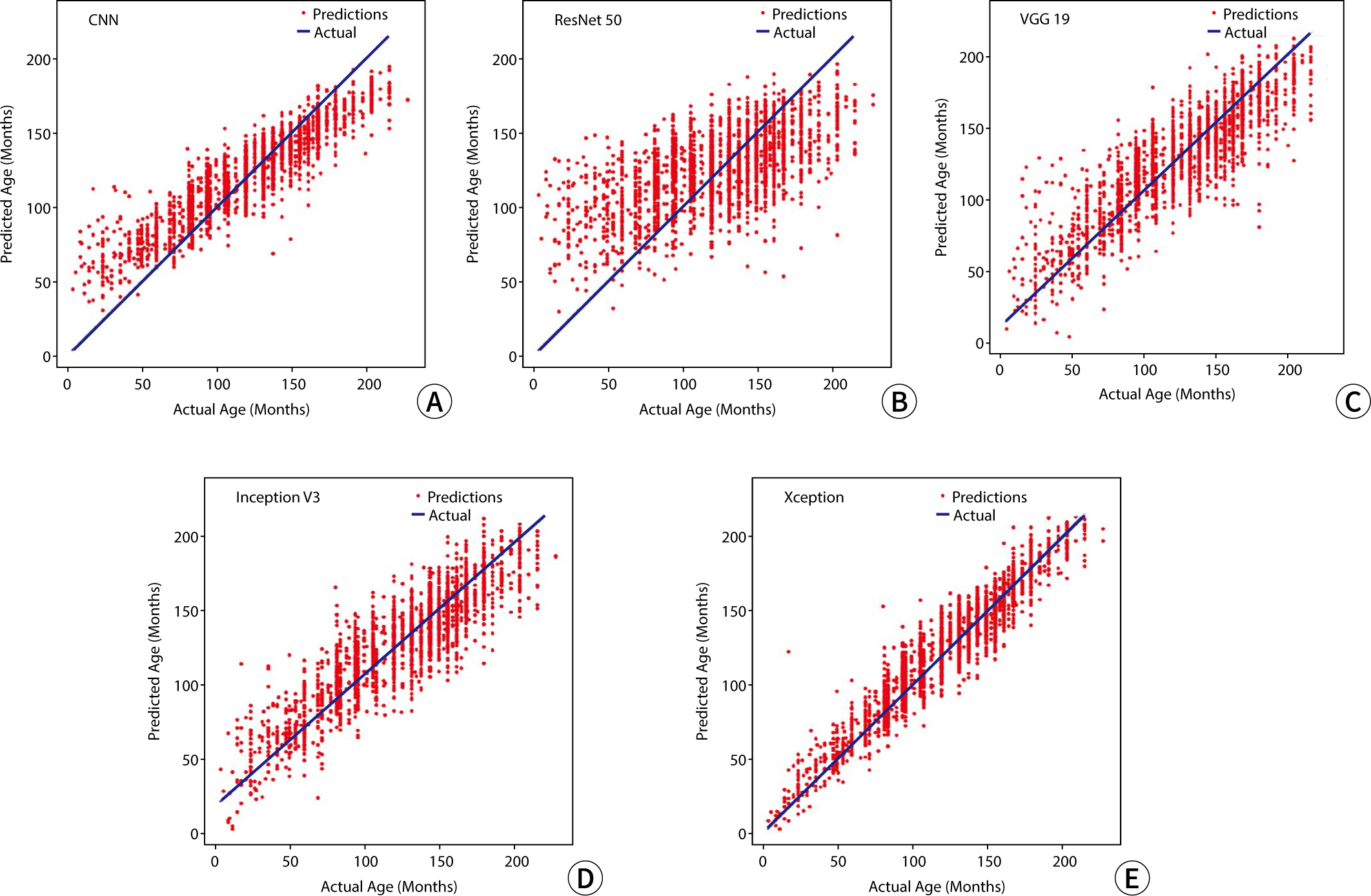
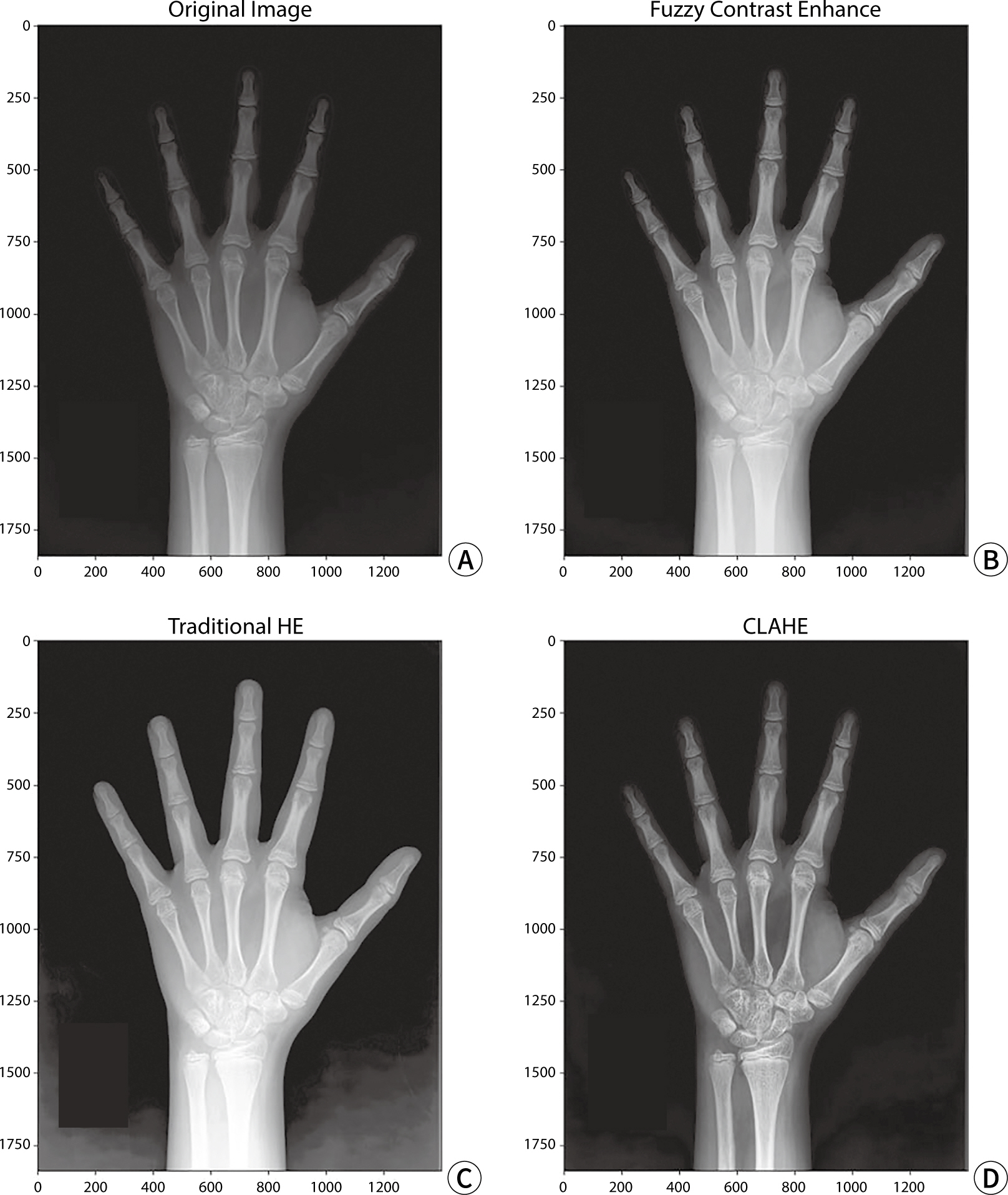
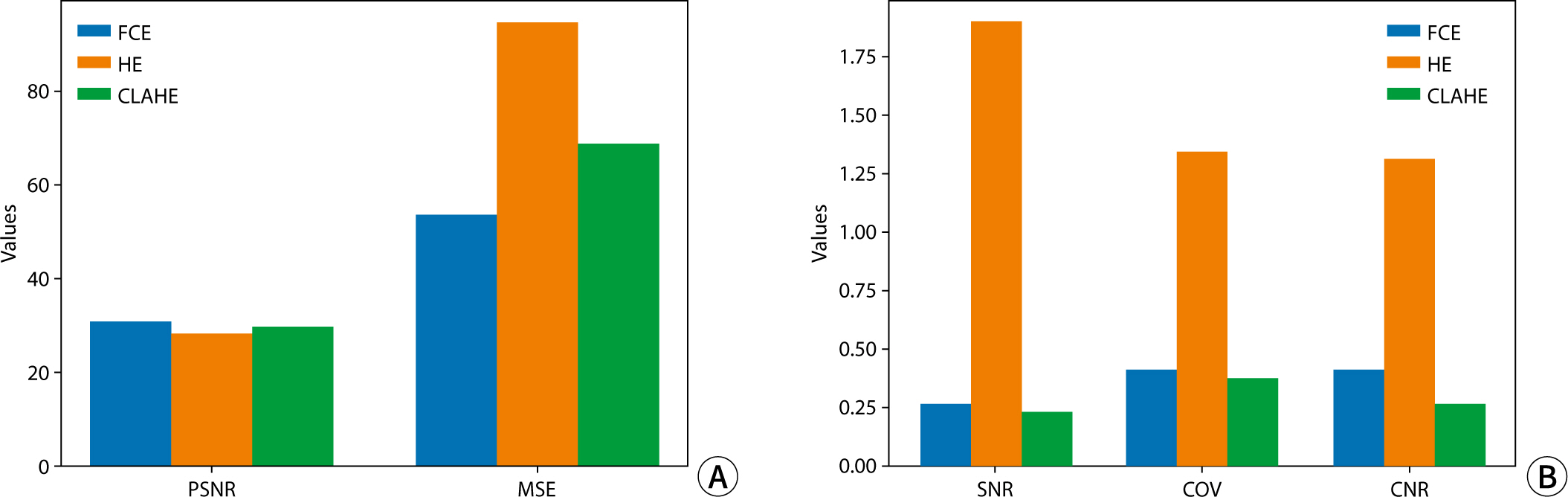
The study employed a variety of deep learning models and contrast conversion techniques to predict bone age. The training dataset consisted of pediatric left-hand X-ray images, each annotated with bone age and sex information. Deep learning models, including a convolutional neural network , Residual Network 50 , Visual Geometry Group 19, Inception V3, and Xception were trained and assessed using the mean absolute error (MAE). For the test data, contrast conversion techniques including fuzzy contrast enhancement, contrast limited adaptive histogram equalization (HE) , and HE were implemented. The quality of the images was evaluated using peak signal-to-noise ratio (SNR), mean squared error, SNR, coefficient of variation, and contrast-to-noise ratio metrics. The bone age prediction results using the test data were evaluated based on the MAE and root mean square error, and the t-test was performed.
Results
The Xception model showed the best performance (MAE=41.12). HE exhibited superior image quality, with higher SNR and coefficient of variation values than other methods. Additionally, HE demonstrated the highest contrast among the techniques assessed, with a contrast-to-noise ratio value of 1.29. Improvements in bone age prediction resulted in a decline in MAE from 2.11 to 0.24, along with a decrease in root mean square error from 0.21 to 0.02.
Conclusion
This study demonstrates that preprocessing the data before model training does not significantly affect the performance of bone age prediction when comparing contrast-converted images with original images.
Keyword
Figure
Reference
-
References
1. Cavallo F, Mohn A, Chiarelli F, Giannini C. Evaluation of bone age in children: a mini-review. Frontiers Pediatr. 2021; 9:580314. DOI: 10.3389/fped.2021.580314. PMID: 33777857. PMCID: PMC7994346.2. Wang S, Wang X, Shen Y, He B, Zhao X, Cheung PWH, et al. An ensemble-based densely-connected deep learning system for assessment of skeletal maturity. IEEE Trans Syst Man Cybern Syst. 2020; 52(1):426–437. DOI: 10.1109/TSMC.2020.2997852.3. Ferrillo M, Curci C, Roccuzzo A, Migliario M, Invernizzi M, de Sire A. Reliability of cervical vertebral maturation compared to hand-wrist for skeletal maturation assessment in growing subjects: a systematic review. J Back Musculoskelet Rehabil. 2021; 34(6):925–936. DOI: 10.3233/BMR-210003. PMID: 33998532.4. Satoh M, Hasegawa Y. Factors affecting prepubertal and pubertal bone age progression. Front Endocrinol. 2022; 13:967711. DOI: 10.3389/fendo.2022.967711. PMID: 36072933. PMCID: PMC9441639.5. Ahn KS, Bae B, Jang WY, Lee JH, Oh S, Kim BH, et al. Assessment of rapidly advancing bone age during puberty on elbow radiographs using a deep neural network model. Eur Radiol. 2021; 31(12):8947–8955. DOI: 10.1007/s00330-021-08096-1. PMID: 34115194.6. Maratova K, Zemkova D, Sedlak P, Pavlikova M, Amaratunga SA, Krasnicanova H, et al. A comprehensive validation study of the latest version of BoneXpert on a large cohort of Caucasian children and adolescents. Front Endocrinol. 2023; 14:1130580. DOI: 10.3389/fendo.2023.1130580. PMID: 37033216. PMCID: PMC10079872.7. Son SJ, Song Y, Kim N, Do Y, Kwak N, Lee MS, et al. TW3-based fully automated bone age assessment system using deep neural networks. IEEE Access. 2019; 7:33346–33358. DOI: 10.1109/ACCESS.2019.2903131.8. Gilsanz V, Ratib O. Hand bone age: a digital atlas of skeletal maturity. Berlin: Springer;2005.9. Kim PH, Yoon HM, Kim JR, Hwang JY, Choi JH, Hwang J, et al. Bone age assessment using artificial intelligence in Korean pediatric population: a comparison of deep-learning models trained with healthy chronological and Greulich-Pyle ages as labels. Korean J Radiol. 2023; 24(11):1151–1163. DOI: 10.3348/kjr.2023.0092. PMID: 37899524. PMCID: PMC10613838.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Bone Age Estimation and Prediction of Final Adult Height Using Deep Learning
- Deep Learning in Dental Radiographic Imaging
- Development of an Optimized Deep Learning Model for Medical Imaging
- Statistics and Deep Belief Network-Based Cardiovascular Risk Prediction
- Development of a deep learning model for predicting critical events in a pediatric intensive care unit