J Stroke.
2024 May;26(2):312-320. 10.5853/jos.2023.03426.
A Multimodal Ensemble Deep Learning Model for Functional Outcome Prognosis of Stroke Patients
- Affiliations
-
- 1Department of Neurology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
- 2Department of Neurology, Kyung Hee University Medical Center, Seoul, Korea
- 3Department of Neurology, Pusan National University Hospital, Busan, Korea
- 4Department of Neurology, Yeungnam University Medical Center, Daegu, Korea
- 5Department of Neurology, Dong-A University Hospital, Busan, Korea
- 6Department of Neurology, Chonnam National University Hospital, Gwangju, Korea
- 7Department of Neurology, Hallym University Sacred Heart Hospital, Anyang, Korea
- 8Department of Neurology, Korea University Ansan Hospital, Ansan, Korea
- 9Department of Neurology, Chosun University Hospital, Gwangju, Korea
- 10Department of Neurology, Dongguk University Ilsan Hospital, Ilsan, Korea
- 11Department of Neurology, Hallym University Kangdong Sacred Heart Hospital, Seoul, Korea
- 12Department of Neurology, Inje University Ilsan Paik Hospital, Ilsan, Korea
- 13Department of Neurology, Keimyung University Medical Center, Daegu, Korea
- 14Department of Neurology, Pusan National University Yangsan Hospital, Yangsan, Korea
- KMID: 2556051
- DOI: http://doi.org/10.5853/jos.2023.03426
Abstract
- Background and Purpose
The accurate prediction of functional outcomes in patients with acute ischemic stroke (AIS) is crucial for informed clinical decision-making and optimal resource utilization. As such, this study aimed to construct an ensemble deep learning model that integrates multimodal imaging and clinical data to predict the 90-day functional outcomes after AIS.
Methods
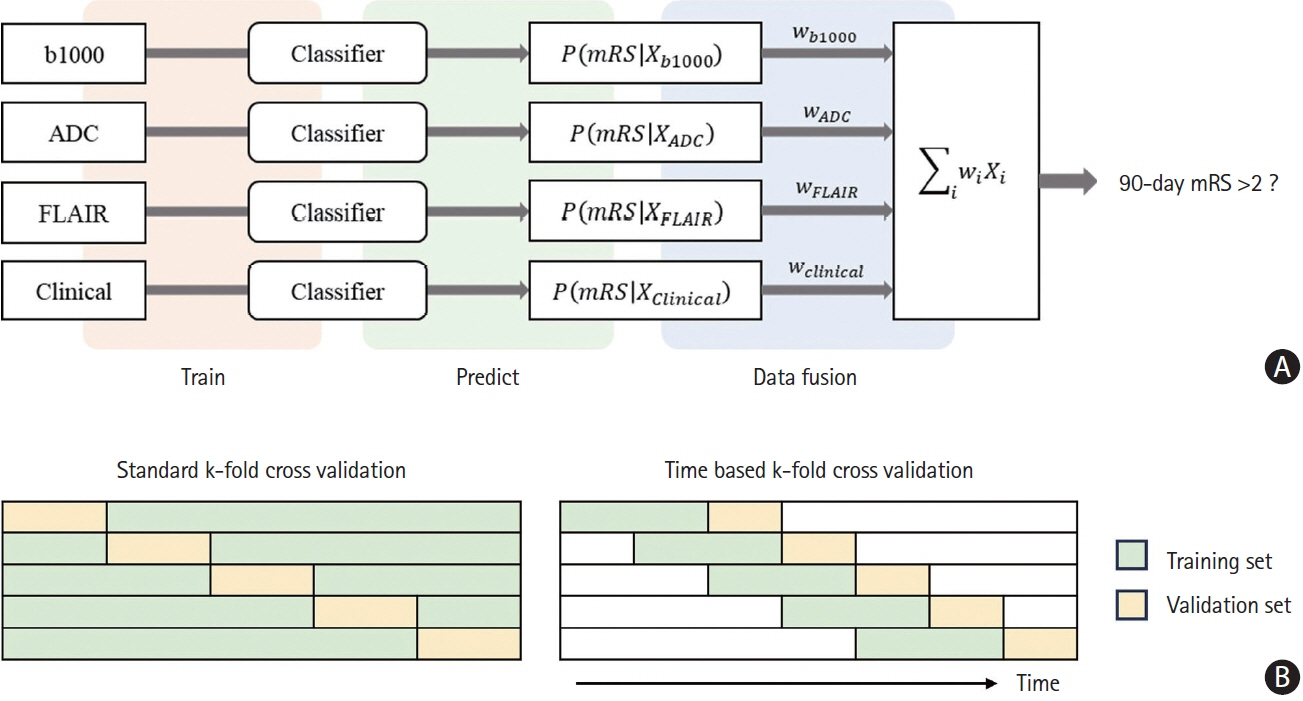
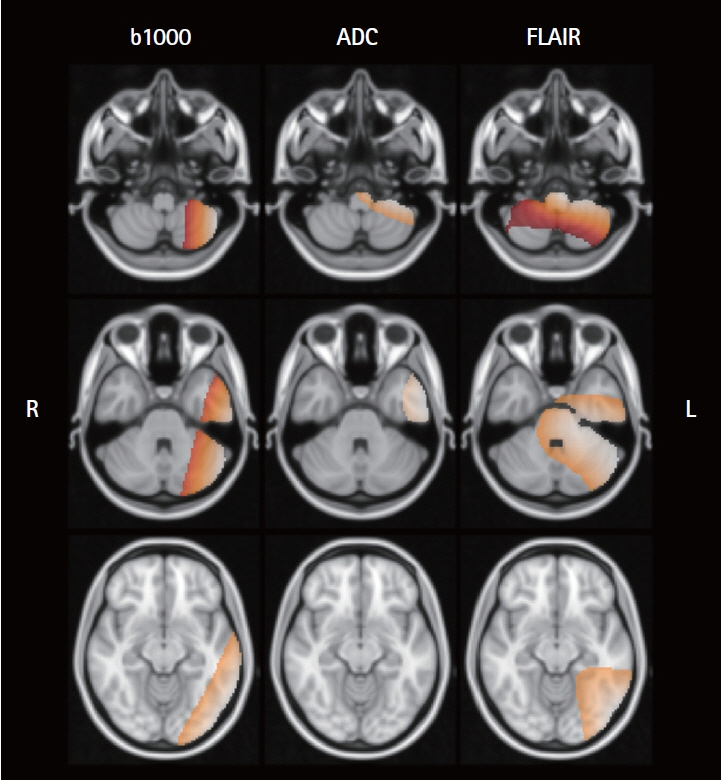
We used data from the Korean Stroke Neuroimaging Initiative database, a prospective multicenter stroke registry to construct an ensemble model integrated individual 3D convolutional neural networks for diffusion-weighted imaging and fluid-attenuated inversion recovery (FLAIR), along with a deep neural network for clinical data, to predict 90-day functional independence after AIS using a modified Rankin Scale (mRS) of 3–6. To evaluate the performance of the ensemble model, we compared the area under the curve (AUC) of the proposed method with that of individual models trained on each modality to identify patients with AIS with an mRS score of 3–6.
Results
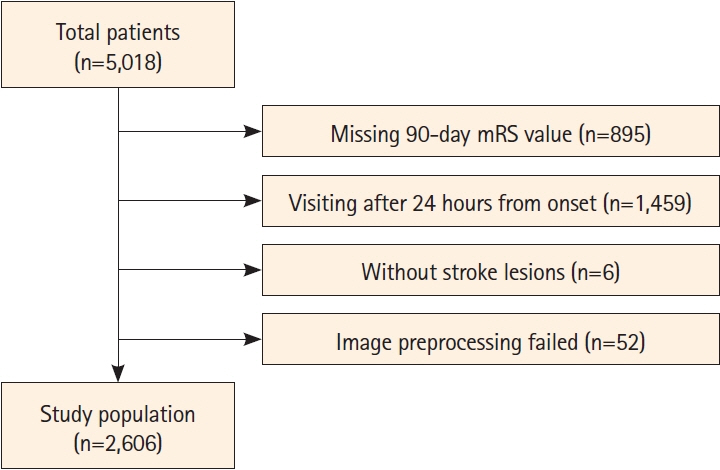
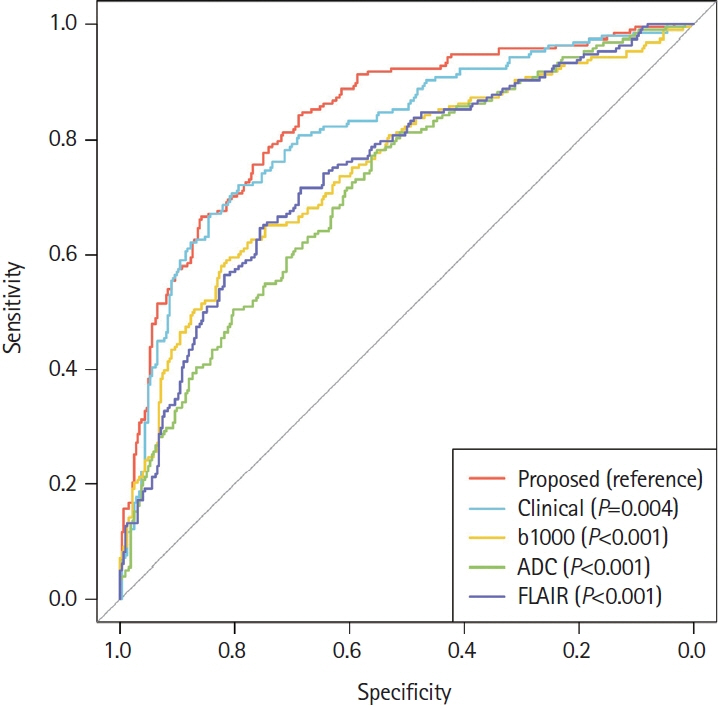
Of the 2,606 patients with AIS, 993 (38.1%) achieved an mRS score of 3–6 at 90 days post-stroke. Our model achieved AUC values of 0.830 (standard cross-validation [CV]) and 0.779 (time-based CV), which significantly outperformed the other models relying on single modalities: b-value of 1,000 s/mm2 (P<0.001), apparent diffusion coefficient map (P<0.001), FLAIR (P<0.001), and clinical data (P=0.004).
Conclusion
The integration of multimodal imaging and clinical data resulted in superior prediction of the 90-day functional outcomes in AIS patients compared to the use of a single data modality.
Keyword
Figure
Reference
-
References
1. James SL, Abate D, Abate KH, Abay SM, Abbafati C, Abbasi N, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the global burden of disease study 2017. Lancet. 2018; 392:1789–1858.2. Langhammer B, Sunnerhagen KS, Lundgren-Nilsson Å, Sällström S, Becker F, Stanghelle JK. Factors enhancing activities of daily living after stroke in specialized rehabilitation: an observational multicenter study within the Sunnaas International Network. Eur J Phys Rehabil Med. 2017; 53:725–734.
Article3. Kugler C, Altenhöner T, Lochner P, Ferbert A. Does age influence early recovery from ischemic stroke? A study from the Hessian Stroke Data Bank. J Neurol. 2003; 250:676–681.4. Venema E, Mulder MJHL, Roozenbeek B, Broderick JP, Yeatts SD, Khatri P, et al. Selection of patients for intra-arterial treatment for acute ischaemic stroke: development and validation of a clinical decision tool in two randomised trials. BMJ. 2017; 357:j1710.5. Paker N, Bugˇdaycı D, Tekdös¸ D, Kaya B, Dere C. Impact of cognitive impairment on functional outcome in stroke. Stroke Res Treat. 2010; 2010:652612.
Article6. Molina CA, Alexandrov AV, Demchuk AM, Saqqur M, Uchino K, Alvarez-Sabín J. Improving the predictive accuracy of recanalization on stroke outcome in patients treated with tissue plasminogen activator. Stroke. 2004; 35:151–156.
Article7. Nichols-Larsen DS, Clark PC, Zeringue A, Greenspan A, Blanton S. Factors influencing stroke survivors’ quality of life during subacute recovery. Stroke. 2005; 36:1480–1484.
Article8. Saposnik G, Guzik AK, Reeves M, Ovbiagele B, Johnston SC. Stroke prognostication using age and NIH stroke scale: SPAN100. Neurology. 2013; 80:21–28.
Article9. Boers AMM, Jansen IGH, Beenen LFM, Devlin TG, San Roman L, Heo JH, et al. Association of follow-up infarct volume with functional outcome in acute ischemic stroke: a pooled analysis of seven randomized trials. J Neurointerv Surg. 2018; 10:1137–1142.10. Laredo C, Zhao Y, Rudilosso S, Renú A, Pariente JC, Chamorro Á, et al. Prognostic significance of infarct size and location: the case of insular stroke. Sci Rep. 2018; 8:9498.
Article11. Kim YD, Choi HY, Jung YH, Yoo J, Nam HS, Song D, et al. The ischemic stroke predictive risk score predicts early neurological deterioration. J Stroke Cerebrovasc Dis. 2016; 25:819–824.
Article12. Ntaios G, Faouzi M, Ferrari J, Lang W, Vemmos K, Michel P. An integer-based score to predict functional outcome in acute ischemic stroke: the ASTRAL score. Neurology. 2012; 78:1916–1922.
Article13. Jang SK, Chang JY, Lee JS, Lee EJ, Kim YH, Han JH, et al. Reliability and clinical utility of machine learning to predict stroke prognosis: comparison with logistic regression. J Stroke. 2020; 22:403–406.
Article14. Heo J, Yoon JG, Park H, Kim YD, Nam HS, Heo JH. Machine learning-based model for prediction of outcomes in acute stroke. Stroke. 2019; 50:1263–1265.15. van Os HJA, Ramos LA, Hilbert A, van Leeuwen M, van Walderveen MAA, Kruyt ND, et al. Predicting outcome of endovascular treatment for acute ischemic stroke: potential value of machine learning algorithms. Front Neurol. 2018; 9:784.
Article16. Ramos LA, Kappelhof M, van Os HJA, Chalos V, Van Kranendonk K, Kruyt ND, et al. Predicting poor outcome before endovascular treatment in patients with acute ischemic stroke. Front Neurol. 2020; 11:580957.
Article17. Alaka SA, Menon BK, Brobbey A, Williamson T, Goyal M, Demchuk AM, et al. Functional outcome prediction in ischemic stroke: a comparison of machine learning algorithms and regression models. Front Neurol. 2020; 11:889.
Article18. Hilbert A, Ramos LA, van Os HJA, Olabarriaga SD, Tolhuisen ML, Wermer MJH, et al. Data-efficient deep learning of radiological image data for outcome prediction after endovascular treatment of patients with acute ischemic stroke. Comput Biol Med. 2019; 115:103516.
Article19. Samak ZA, Clatworthy P, Mirmehdi M. Prediction of thrombectomy functional outcomes using multimodal data. In: Papiez˙ B, Namburete A, Yaqub M, Noble J. Medical image understanding and analysis. MIUA 2020. Communications in computer and information science (vol 1248). Cham: Springer, 2020;267-279.20. Hatami N, Cho TH, Mechtouff L, Eker OF, Rousseau D, Frindel C. CNN-LSTM based multimodal MRI and clinical data fusion for predicting functional outcome in stroke patients. Annu Int Conf IEEE Eng Med Biol Soc. 2022; 2022:3430–3434.
Article21. Moulton E, Valabregue R, Piotin M, Marnat G, Saleme S, Lapergue B, et al. Interpretable deep learning for the prognosis of long-term functional outcome post-stroke using acute diffusion weighted imaging. J Cereb Blood Flow Metab. 2023; 43:198–209.
Article22. van Swieten JC, Koudstaal PJ, Visser MC, Schouten HJ, van Gijn J. Interobserver agreement for the assessment of handicap in stroke patients. Stroke. 1988; 19:604–607.
Article23. Sulter G, Steen C, De Keyser J. Use of the Barthel index and modified Rankin scale in acute stroke trials. Stroke. 1999; 30:1538–1541.
Article24. Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010; 29:1310–1320.
Article25. Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008; 12:26–41.
Article26. Adams HP Jr, Bendixen BH, Kappelle LJ, Biller J, Love BB, Gordon DL, et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993; 24:35–41.
Article27. Kingma DP, Ba J. Adam: a method for stochastic optimization. arXiv [Preprint]. 2014 [accessed 2023 April 18]. Available from: https://doi.org/10.48550/arXiv.1412.6980.
Article28. Xie S, Girshick R, Dollár P, Tu Z, He K. Aggregated residual transformations for deep neural networks. Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017 Jul 21-26; Honolulu, HI, USA. New York: IEEE, 2017. p.5987-5995.29. Woo S, Park J, Lee JY, Kweon IS. CBAM: convolutional block attention module. In: Ferrari V, Hebert M, Sminchisescu C, Weiss Y. Computer vision – ECCV 2018. Lecture notes in computer science (vol 11211). Cham: Springer;2018. p. 3–19.30. Liu L, Jiang H, He P, Chen W, Liu X, Gao J, et al. On the variance of the adaptive learning rate and beyond. arXiv [Preprint]. 2019 [accessed 2023 March 7]. Available from: https://doi.org/10.48550/arxiv.1908.03265.
Article31. Loshchilov I, Hutter F. SGDR: stochastic gradient descent with warm restarts. arXiv [Preprint]. 2016 [accessed 2023 March 7]. Available from: https://doi.org/10.48550/arxiv.1608.03983.
Article32. Cubuk ED, Zoph B, Shlens J, Le QV. RandAugment: practical automated data augmentation with a reduced search space. Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW); 2020 Jun 14-19; Seattle, WA, USA. New York: IEEE, 2020. p.3008-3017.33. Lin TY, Goyal P, Girshick R, He K, Dollar P. Focal loss for dense object detection. IEEE Trans Pattern Anal Mach Intell. 2020; 42:318–327.34. Lundberg SM, Lee SI. A unified approach to interpreting model predictions. In: Guyon I, Luxburg U Von, Bengio S, Wallach H, Fergus R, Vishwanathan S, et al. editors, eds. 31st Conference on Neural Information Processing Systems (NIPS 2017); 2017 Dec 4-9; Long Beach, CA, USA. San Diego, CA: Advances in Neural Information Processing Systems; 2017. p. 4765-4774.35. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: visual explanations from deep networks via gradient-based localization. Int J Comput Vis. 2020; 128:336–359.
Article36. Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, et al. Tensorflow: a system for large-scale machine learning. Proceedings of the 12th USENIX Conference on Operating Systems Design and Implementation; 2016 Nov 2–4; Savannah, GA, USA. Berkeley: USENIX Association; 2016. p. 265-283.37. Bradski G. The opencv library. Dr Dobb’s Journal: Software Tools for the Professional Programmer. 2000; 25:120–123.38. van der Walt S, Schönberger JL, Nunez-Iglesias J, Boulogne F, Warner JD, Yager N, et al. Scikit-image: image processing in Python. PeerJ. 2014; 2:e453.
Article39. Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011; 12:2825–2830.40. Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. FSL. Neuroimage. 2012; 62:782–790.
Article41. Corso G, Bottacchi E, Tosi P, Caligiana L, Lia C, Veronese Morosini M, et al. Outcome predictors in first-ever ischemic stroke patients: a population-based study. Int Sch Res Notices. 2014; 2014:904647.
Article42. Turhan B. On the dataset shift problem in software engineering prediction models. Empir Software Eng. 2012; 17:62–74.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Stacking Ensemble Technique for Classifying Breast Cancer
- Management Architecture With Multi-modal Ensemble AI Models for Worker Safety
- Development of an Optimized Deep Learning Model for Medical Imaging
- Deep Learning Model-Based Detection of Anemia from Conjunctiva Images
- Development of Early-Stage Stroke Diagnosis System for the Elderly Neurogenic Bladder Prevention