Ann Lab Med.
2024 May;44(3):294-298. 10.3343/alm.2023.0267.
Optimization of a Protocol for Isolating Cell-free DNA From Cerebrospinal Fluid
- Affiliations
-
- 1Department of Interdisciplinary Program in Biomedical Science, Graduate School, Soonchunhyang University, Asan, Korea
- 2Department of Neurosurgery, Soonchunhyang University Seoul Hospital, Seoul, Korea
- 3Department of Laboratory Medicine, Soonchunhyang University Seoul Hospital, Seoul, Korea
- 4Department of Neurosurgery, Soonchunhyang University Cheonan Hospital, Cheonan, Korea
- KMID: 2555708
- DOI: http://doi.org/10.3343/alm.2023.0267
Abstract
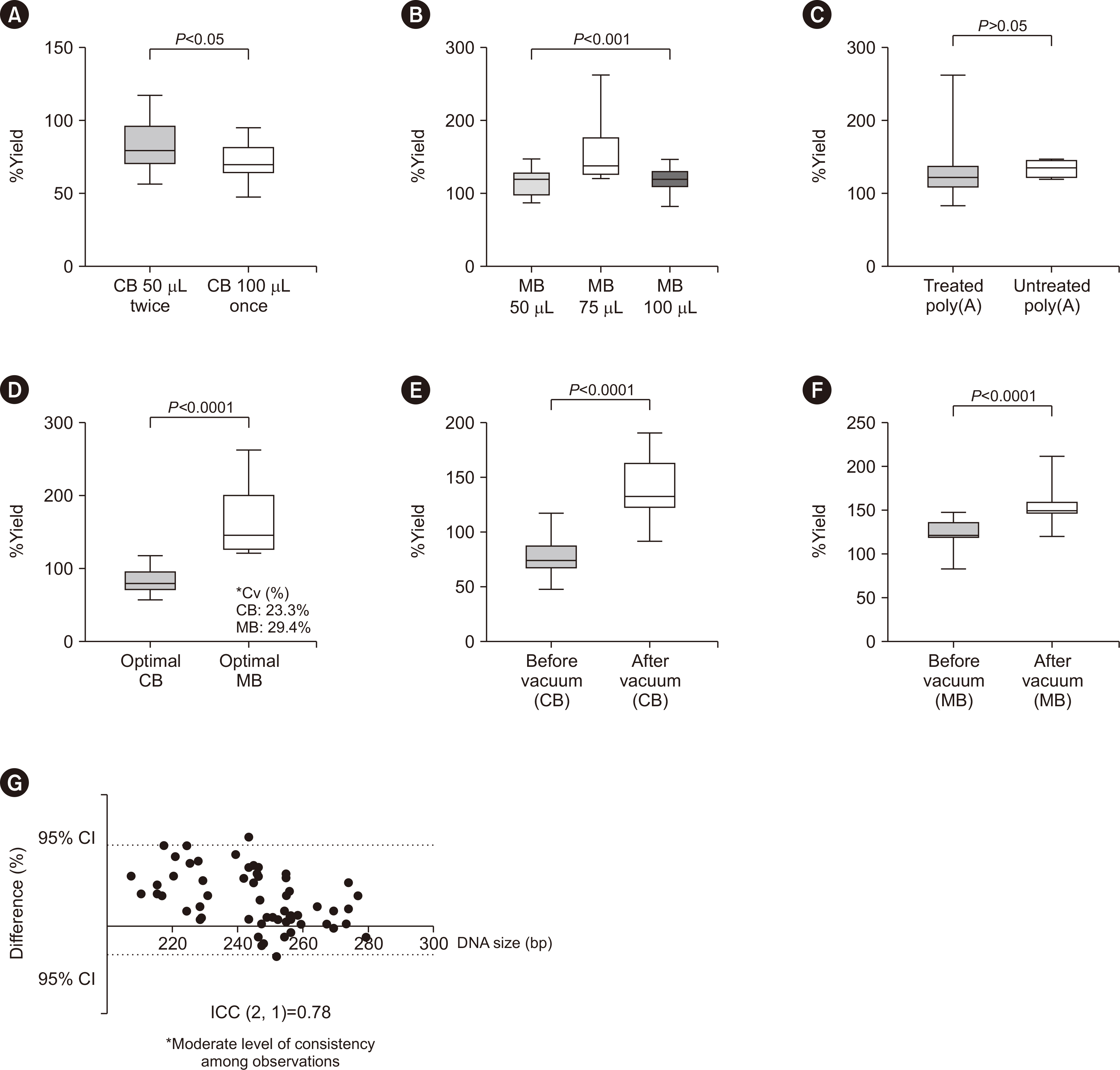
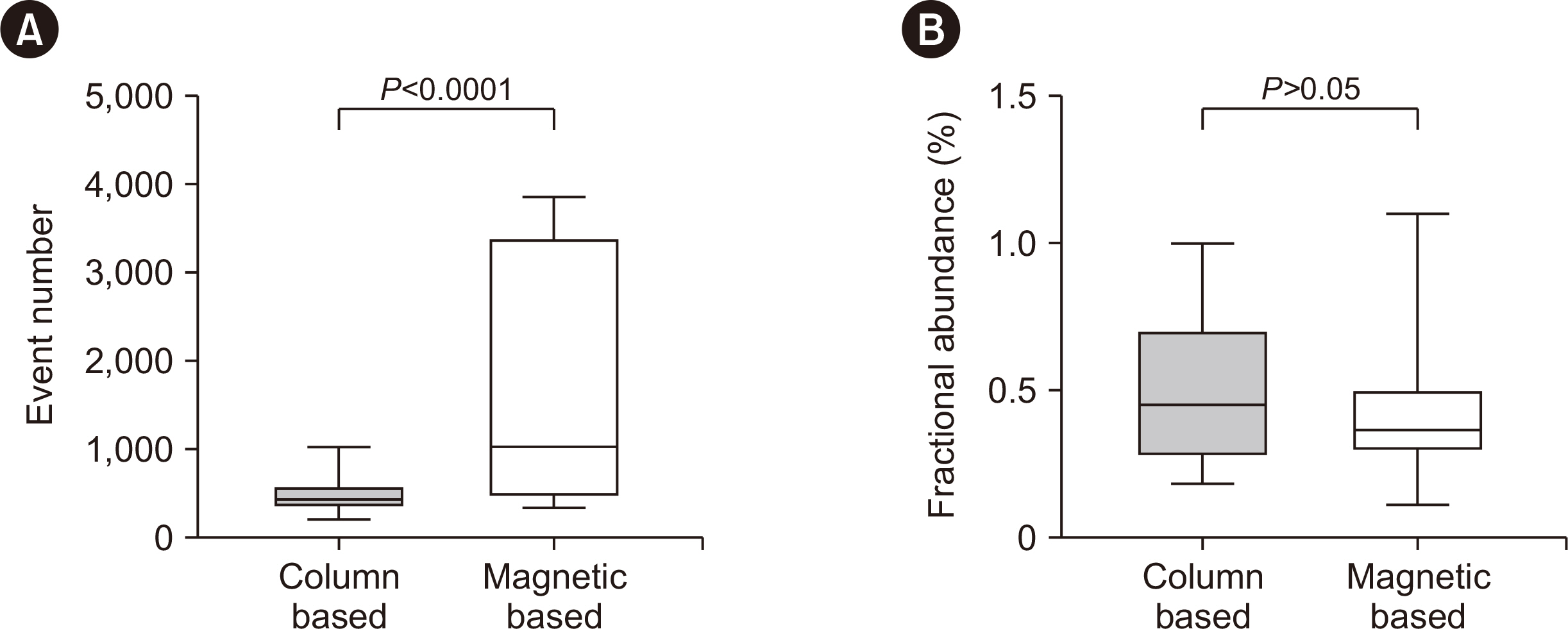
- A standardized protocol for the isolation of cell-free DNA (cfDNA) from cerebrospinal fluid (CSF) is lacking. Therefore, we established a cfDNA isolation protocol optimized for clinical CSF specimens, integrating acceptable modifications and using artificial CSF generated from remnant CSF spiked with reference cell-free tumor DNA (ctDNA). We compared the isolation yields of in vitro diagnostic (IVD)-certified column-based (CB) and magnetic beadbased (MB) isolation. Furthermore, we modified both methods, including pre- and postelution steps. To confirm ctDNA integrity and quantify the variant allele frequency after isolation, we performed droplet digital PCR (ddPCR) targeting IDH1 R132C in the reference ctDNA. MB isolation had a higher yield than CB isolation (P < 0.0001), and post-isolation vacuum increased the final concentration in both methods, with little effect on cfDNA integrity. Our study provides a protocol to maximize CSF-ctDNA concentrations in IVD testing and future studies.
Keyword
Figure
Reference
-
References
1. Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. 2021; The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 23:1231–51. DOI: 10.1093/neuonc/noab106. PMID: 34185076. PMCID: PMC8328013.2. Miller AM, Shah RH, Pentsova EI, Pourmaleki M, Briggs S, Distefano N, et al. 2019; Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature. 565:654–8. DOI: 10.1038/s41586-019-0882-3. PMID: 30675060. PMCID: PMC6457907.3. Teunissen CE, Verheul C, Willemse EAJ. Deisenhammer F, Teunissen CE, editors. 2017. The use of cerebrospinal fluid in biomarker studies. Handbook of Clinical Neurology. Elsevier;p. 3–20. DOI: 10.1016/B978-0-12-804279-3.00001-0. PMID: 29110777.4. Zhong Y, Fan Q, Zhou Z, Wang Y, He K, Lu J. 2020; Plasma cfDNA as a potential biomarker to evaluate the efficacy of chemotherapy in gastric cancer. Cancer Manag Res. 12:3099–106. DOI: 10.2147/CMAR.S243320. PMID: 32440208. PMCID: PMC7211302.5. Kim S, Baldassari S, Sim NS, Chipaux M, Dorfmüller G, Kim DS, et al. 2021; Detection of brain somatic mutations in cerebrospinal fluid from refractory epilepsy patients. Ann Neurol. 89:1248–52. DOI: 10.1002/ana.26080. PMID: 33834539.6. Sheng Z, Yu J, Deng K, Andrade-Barazarte H, Zemmar A, Li S, et al. 2021; Characterizing the genomic landscape of brain glioma with circulating tumor DNA from tumor in situ fluid. Front Oncol. 11:584988. DOI: 10.3389/fonc.2021.584988. PMID: 33868989. PMCID: PMC8045748.7. Bronkhorst AJ, Ungerer V, Holdenrieder S. 2020; Comparison of methods for the isolation of cell-free DNA from cell culture supernatant. Tumour Biol. 42:1010428320916314. DOI: 10.1177/1010428320916314. PMID: 32338581.8. Katevatis C, Fan A, Klapperich CM. 2017; Low concentration DNA extraction and recovery using a silica solid phase. PLoS One. 12:e0176848. DOI: 10.1371/journal.pone.0176848. PMID: 28475611. PMCID: PMC5419563.9. Desneux J, Pourcher AM. 2014; Comparison of DNA extraction kits and modification of DNA elution procedure for the quantitation of subdominant bacteria from piggery effluents with real-time PCR. Microbiologyopen. 3:437–45. DOI: 10.1002/mbo3.178. PMID: 24838631. PMCID: PMC4287173.10. Bronkhorst AJ, Aucamp J, Pretorius PJ. 2015; Cell-free DNA: preanalytical variables. Clin Chim Acta. 450:243–53. DOI: 10.1016/j.cca.2015.08.028. PMID: 26341895.11. Shin S, Woo HI, Kim J-W, Kim Y, Lee KA. 2022; Clinical practice guidelines for pre-analytical procedures of plasma epidermal growth factor receptor variant testing. Ann Lab Med. 42:141–9. DOI: 10.3343/alm.2022.42.2.141. PMID: 34635607. PMCID: PMC8548242.12. Cho SM, Lee HS, Jeon S, Kim Y, Kong SY, Lee KA. 2023; Cost-effectiveness analysis of three diagnostic strategies for the detection of EGFR mutation in advanced non-small cell lung cancer. Ann Lab Med. 43:605–13. DOI: 10.3343/alm.2023.43.6.605. PMID: 37387493. PMCID: PMC10345179.13. Ling J, Wang H, Zhan S, Zhang D, Lai M, Zhu Y. 2012; Optimization of genomic DNA extraction with magnetic bead- based semi-automatic system. Zhejiang Univ (Med Sci). 41:320–6.14. Beránek M, Sirák I, Vošmik M, Petera J, Drastíková M, Palička V. 2016; Carrier molecules and extraction of circulating tumor DNA for next generation sequencing in colorectal cancer. Acta Medica (Hradec Kralove). 59:54–8. DOI: 10.14712/18059694.2016.54. PMID: 27526306.15. Sanchez I, Betsou F, Mathieson W. 2019; Does vacuum centrifugal concentration reduce yield or quality of nucleic acids extracted from FFPE biospecimens? Anal Biochem. 566:16–9. DOI: 10.1016/j.ab.2018.10.020. PMID: 30343041.16. Leest PV, Boonstra PA, Elst AT, Kempen LCV, Tibbesma M, Koopmans J. Comparison of circulating cell-free DNA extraction methods for downstream analysis in cancer patients. Cancers (Basel). 2020; 12:DOI: 10.3390/cancers12051222. PMID: 32414097. PMCID: PMC7281769.17. Pedini P, Graiet H, Laget L, Filosa L, Chatron J, Cherouat N, et al. 2021; Qualitative and quantitative comparison of cell-free DNA and cell-free fetal DNA isolation by four (semi-) automated extraction methods: impact in two clinical applications: chimerism quantification and noninvasive prenatal diagnosis. J Translat Med. 19:15. DOI: 10.1186/s12967-020-02671-8. PMID: 33407582. PMCID: PMC7788686.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Traumatic Cerebrospinal Fluid Rhinorrhea: Successful Closure under the Surgical Microscope
- Surgical Repair of Cerebrospinal Fluid Rhinorrhea with Mucoperichondrial Free Graft
- A Case of Spontaneous Intracranial Hypotension: Detection of Cerebrospinal Fluid Leakage by Early Dynamic Radionuclide Cisternography
- Endoscopic treatment of iatrogenic cerebrospinal fluid rhinorrhea
- Surgical management of iatrogenic cerebrospinal fluid rhinorrhea