J Korean Neurosurg Soc.
2024 May;67(3):364-375. 10.3340/jkns.2023.0155.
High Expression of KIFC1 in Glioma Correlates with Poor Prognosis
- Affiliations
-
- 1Department of Neurosurgery, Liaocheng People’s Hospital, Liaocheng, China
- 2Joint Laboratory for Translational Medicine Research, Liaocheng People’s Hospital, Liaocheng, China
- 3Department of Neurosurgery, Yantaishan Hospital Affiliated to Binzhou Medical University, Yantai, China
- KMID: 2554874
- DOI: http://doi.org/10.3340/jkns.2023.0155
Abstract
Objective
: Kinesin family member C1 (KIFC1), a non-essential kinesin-like motor protein, has been found to serve a crucial role in supernumerary centrosome clustering and the progression of several human cancer types. However, the role of KIFC1 in glioma has been rarely reported. Thus, the present study aimed to investigate the role of KIFC1 in glioma progression.
Methods
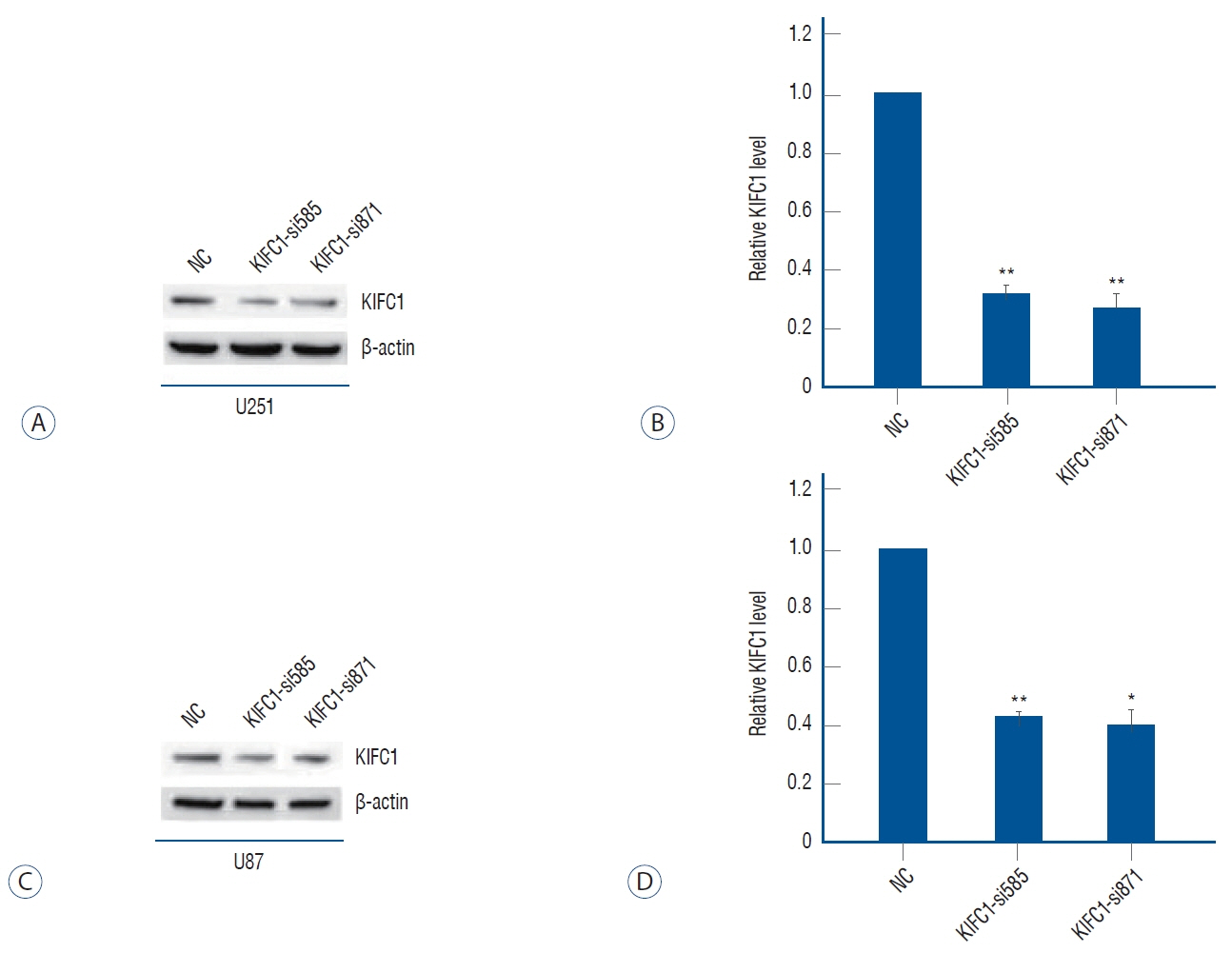
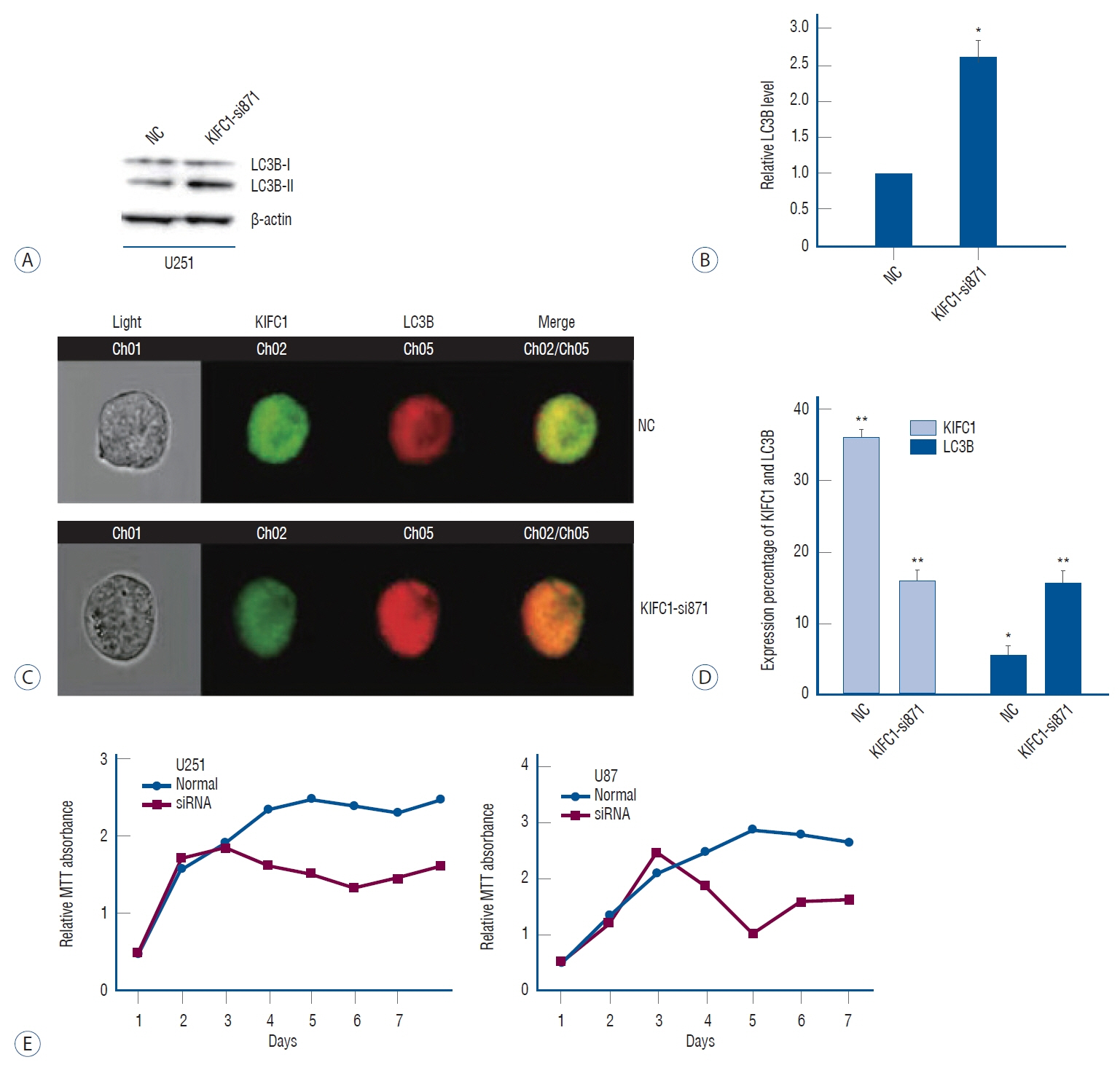
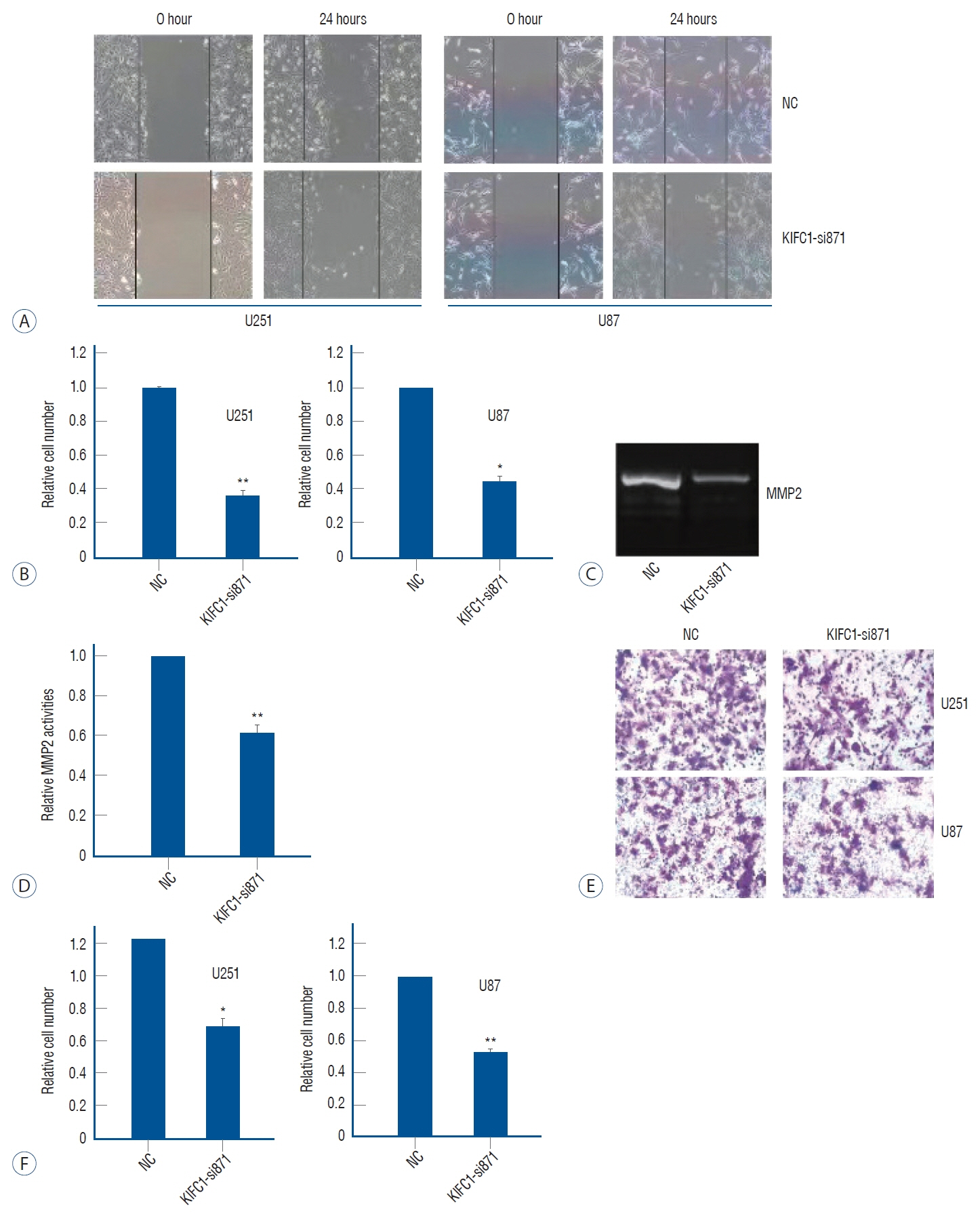
: Online bioinformatics analysis was performed to determine the association between KIFC1 expression and clinical outcomes in glioma. Immunohistochemical staining was conducted to analyze the expression levels of KIFC1 in glioma and normal brain tissues. Furthermore, KIFC1 expression was knocked in the glioma cell lines, U251 and U87MG, and the functional roles of KIFC1 in cell proliferation, invasion and migration were analyzed using cell multiplication, wound healing and Transwell invasion assays, respectively. The autophagic flux and expression levels matrix metalloproteinase-2 (MMP2) were also determined using imaging flow cytometry, western blotting and a gelation zymography assay.
Results
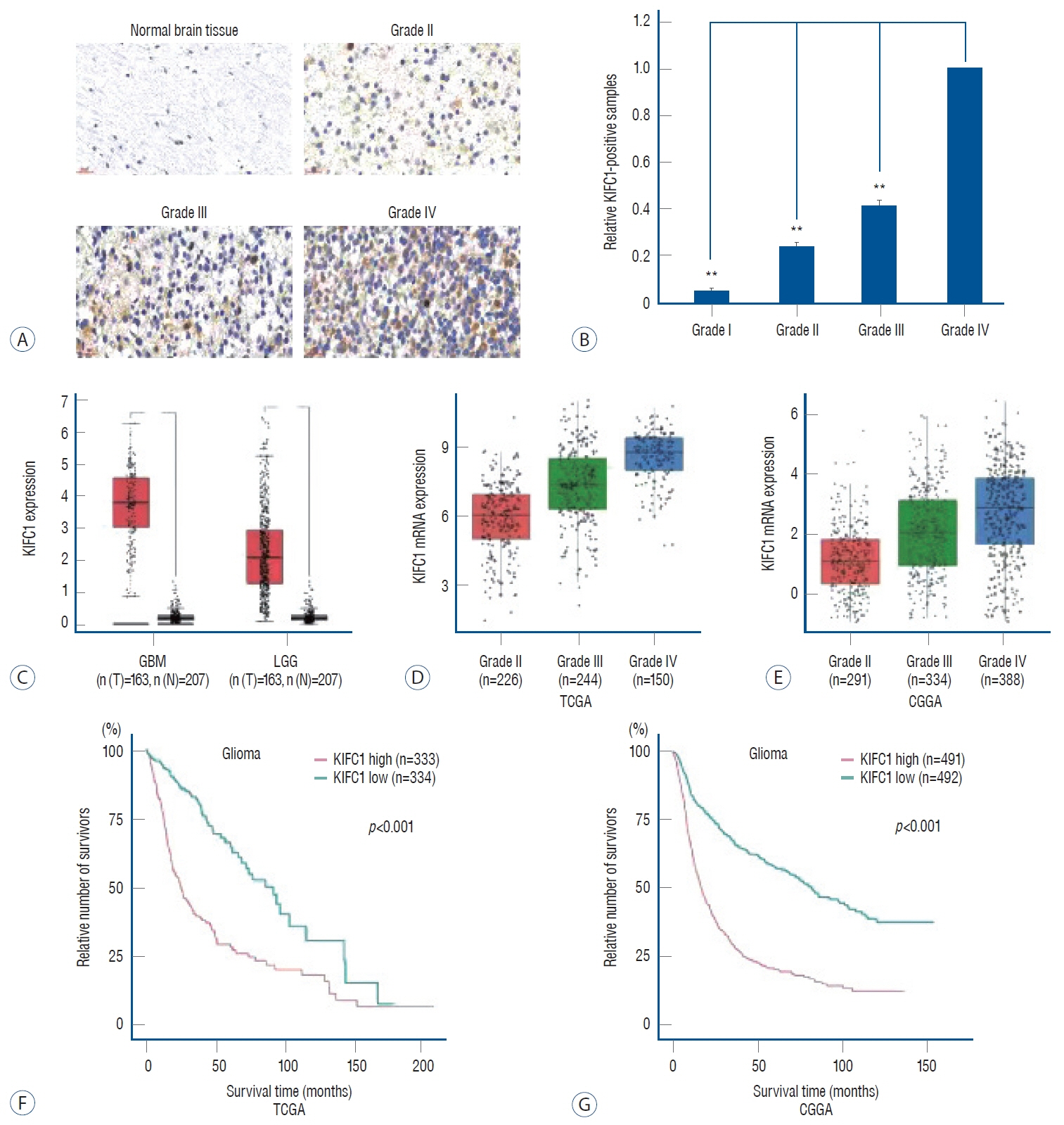
: The results revealed that KIFC1 expression levels were significantly upregulated in glioma tissues compared with normal brain tissues, and the expression levels were positively associated with tumor grade. Patients with glioma with low KIFC1 expression levels had a more favorable prognosis compared with patients with high KIFC1 expression levels. In vitro, KIFC1 knockdown not only inhibited the proliferation, migration and invasion of glioma cells, but also increased the autophagic flux and downregulated the expression levels of MMP2.
Conclusion
: Upregulation of KIFC1 expression may promote glioma progression and KIFC1 may serve as a potential prognostic biomarker and possible therapeutic target for glioma.
Keyword
Figure
Reference
-
References
1. Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 444:756–760. 2006.
Article2. Bowman RL, Wang Q, Carro A, Verhaak RG, Squatrito M. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol. 19:139–141. 2017.
Article3. Conduit PT, Wainman A, Raff JW. Centrosome function and assembly in animal cells. Nat Rev Mol Cell Biol. 16:611–624. 2015.
Article4. Dai J, Bing Z, Zhang Y, Li Q, Niu L, Liang W, et al. Integrated mRNAseq and microRNAseq data analysis for grade III gliomas. Mol Med Rep. 16:7468–7478. 2017.
Article5. Diwanji TP, Engelman A, Snider JW, Mohindra P. Epidemiology, diagnosis, and optimal management of glioma in adolescents and young adults. Adolesc Health Med Ther. 8:99–113. 2017.
Article6. Dong W, Li H, Zhang Y, Yang H, Guo M, Li L, et al. Matrix metalloproteinase 2 promotes cell growth and invasion in colorectal cancer. Acta Biochim Biophys Sin (Shanghai). 43:840–848. 2011.
Article7. Fu X, Zhu Y, Zheng B, Zou Y, Wang C, Wu P, et al. KIFC1, a novel potential prognostic factor and therapeutic target in hepatocellular carcinoma. Int J Oncol. 52:1912–1922. 2018.
Article8. Global Burden of Disease Cancer Collaboration, Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 3:524–548. 2017.9. Grinberg-Rashi H, Ofek E, Perelman M, Skarda J, Yaron P, Hajdúch M, et al. The expression of three genes in primary non-small cell lung cancer is associated with metastatic spread to the brain. Clin Cancer Res. 15:1755–1761. 2009.
Article10. Han J, Wang F, Lan Y, Wang J, Nie C, Liang Y, et al. KIFC1 regulated by miR-532-3p promotes epithelial-to-mesenchymal transition and metastasis of hepatocellular carcinoma via gankyrin/AKT signaling. Oncogene. 38:406–420. 2019.
Article11. Kawai H, Ishii A, Washiya K, Konno T, Kon H, Yamaya C, et al. Estrogen receptor alpha and beta are prognostic factors in non-small cell lung cancer. Clin Cancer Res. 11:5084–5089. 2005.
Article12. Kleylein-Sohn J, Pöllinger B, Ohmer M, Hofmann F, Nigg EA, Hemmings BA, et al. Acentrosomal spindle organization renders cancer cells dependent on the kinesin HSET. J Cell Sci. 125(Pt 22):5391–5402. 2012.
Article13. Klionsky DJ. Autophagy revisited: a conversation with Christian de Duve. Autophagy. 4:740–743. 2008.
Article14. Levine MS, Bakker B, Boeckx B, Moyett J, Lu J, Vitre B, et al. Centrosome amplification is sufficient to promote spontaneous tumorigenesis in mammals. Dev Cell. 40:313–322.e5. 2017.
Article15. Li Y, Lu W, Chen D, Boohaker RJ, Zhai L, Padmalayam I, et al. KIFC1 is a novel potential therapeutic target for breast cancer. Cancer Biol Ther. 16:1316–1322. 2015.
Article16. Liu Y, Pelletier L. A magic bullet for targeting cancers with supernumerary centrosomes. EMBO J. 38:e101134. 2019.
Article17. Liu Y, Zhan P, Zhou Z, Xing Z, Zhu S, Ma C, et al. The overexpression of KIFC1 was associated with the proliferation and prognosis of non-small cell lung cancer. J Thorac Dis. 8:2911–2923. 2016.
Article18. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 114:97–109. 2007.
Article19. Mittal K, Choi DH, Klimov S, Pawar S, Kaur R, Mitra AK, et al. A centrosome clustering protein, KIFC1, predicts aggressive disease course in serous ovarian adenocarcinomas. J Ovarian Res. 9:17. 2016.
Article20. Pallichankandy S, Rahman A, Thayyullathil F, Galadari S. ROS-dependent activation of autophagy is a critical mechanism for the induction of anti-glioma effect of sanguinarine. Free Radic Biol Med. 89:708–720. 2015.
Article21. Pawar S, Donthamsetty S, Pannu V, Rida P, Ogden A, Bowen N, et al. KIFCI, a novel putative prognostic biomarker for ovarian adenocarcinomas: delineating protein interaction networks and signaling circuitries. J Ovarian Res. 7:53. 2014.
Article22. Rath O, Kozielski F. Kinesins and cancer. Nat Rev Cancer. 12:527–539. 2012.
Article23. Sekino Y, Oue N, Koike Y, Shigematsu Y, Sakamoto N, Sentani K, et al. KIFC1 inhibitor CW069 induces apoptosis and reverses resistance to docetaxel in prostate cancer. J Clin Med. 8:225. 2019.
Article24. Sekino Y, Oue N, Shigematsu Y, Ishikawa A, Sakamoto N, Sentani K, et al. KIFC1 induces resistance to docetaxel and is associated with survival of patients with prostate cancer. Urol Oncol. 35:31.e13–31.e20. 2017.
Article25. Shin SY, Lee KS, Choi YK, Lim HJ, Lee HG, Lim Y, et al. The antipsychotic agent chlorpromazine induces autophagic cell death by inhibiting the Akt/mTOR pathway in human U-87MG glioma cells. Carcinogenesis. 34:2080–2089. 2013.
Article26. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 47(W1):W556–W560. 2019.
Article27. Teng K, Wei S, Zhang C, Chen J, Chen J, Xiao K, et al. KIFC1 is activated by TCF-4 and promotes hepatocellular carcinoma pathogenesis by regulating HMGA1 transcriptional activity. J Exp Clin Cancer Res. 38:329. 2019.
Article28. Webb AH, Gao BT, Goldsmith ZK, Irvine AS, Saleh N, Lee RP, et al. Inhibition of MMP-2 and MMP-9 decreases cellular migration, and angiogenesis in in vitro models of retinoblastoma. BMC Cancer. 17:434. 2017.
Article29. Wen ZP, Zeng WJ, Chen YH, Li H, Wang JY, Cheng Q, et al. Knockdown ATG4C inhibits gliomas progression and promotes temozolomide chemosensitivity by suppressing autophagic flux. J Exp Clin Cancer Res. 38:298. 2019.
Article30. Xiao KH, Teng K, Ye YL, Tan L, Chen MK, Liang HT, et al. Kinesin family member C1 accelerates bladder cancer cell proliferation and induces epithelial-mesenchymal transition via Akt/GSK3β signaling. Cancer Sci. 110:2822–2833. 2019.
Article31. Xiao YX, Yang WX. KIFC1: a promising chemotherapy target for cancer treatment? Oncotarget. 7:48656–48670. 2016.
Article32. Yu Y, Feng YM. The role of kinesin family proteins in tumorigenesis and progression: potential biomarkers and molecular targets for cancer therapy. Cancer. 116:5150–5160. 2010.
Article33. Zhang H, Ma Y, Wang H, Xu L, Yu Y. MMP-2 expression and correlation with pathology and MRI of glioma. Oncol Lett. 17:1826–1832. 2019.
Article34. Zhou K, Zhao J, Qi L, He Y, Xu J, Dai M. Kinesin family member C1 (KIFC1) accelerates proliferation and invasion of endometrial cancer cells through modulating the PI3K/AKT signaling pathway. Technol Cancer Res Treat. 19:1533033820964217. 2020.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- CDK1 promotes the phosphorylation of KIFC1 to regulate the tumorgenicity of endometrial carcinoma
- FoxD2-AS1 is a prognostic factor in glioma and promotes temozolomide resistance in a Oâ¶-methylguanine-DNA methyltransferase-dependent manner
- BIRC5 Is a Potential Biomarker Associated with Immune System Infiltration in Glioma
- High Expression of Ribonucleotide Reductase Subunit M2 Correlates with Poor Prognosis of Hepatocellular Carcinoma
- Overview of CNS Gliomas in Childhood