Ann Pediatr Endocrinol Metab.
2024 Apr;29(2):102-108. 10.6065/apem.2346050.025.
Clinical validation of a deep-learning-based bone age software in healthy Korean children
- Affiliations
-
- 1Department of Pediatrics, Korea University College of Medicine, Seoul, Korea
- 2Department of Radiology, Korea University College of Medicine, Seoul, Korea
- 3Smart Health Care Center, Korea University Guro Hospital, Seoul, Korea
- 4Korea University Guro Hospital-Medical Image Data Center (KUGH-MIDC), Seoul, Korea
- KMID: 2554681
- DOI: http://doi.org/10.6065/apem.2346050.025
Abstract
- Purpose
Bone age (BA) is needed to assess developmental status and growth disorders. We evaluated the clinical performance of a deep-learning-based BA software to estimate the chronological age (CA) of healthy Korean children.
Methods
This retrospective study included 371 healthy children (217 boys, 154 girls), aged between 4 and 17 years, who visited the Department of Pediatrics for health check-ups between January 2017 and December 2018. A total of 553 left-hand radiographs from 371 healthy Korean children were evaluated using a commercial deep-learning-based BA software (BoneAge, Vuno, Seoul, Korea). The clinical performance of the deep learning (DL) software was determined using the concordance rate and Bland-Altman analysis via comparison with the CA.
Results
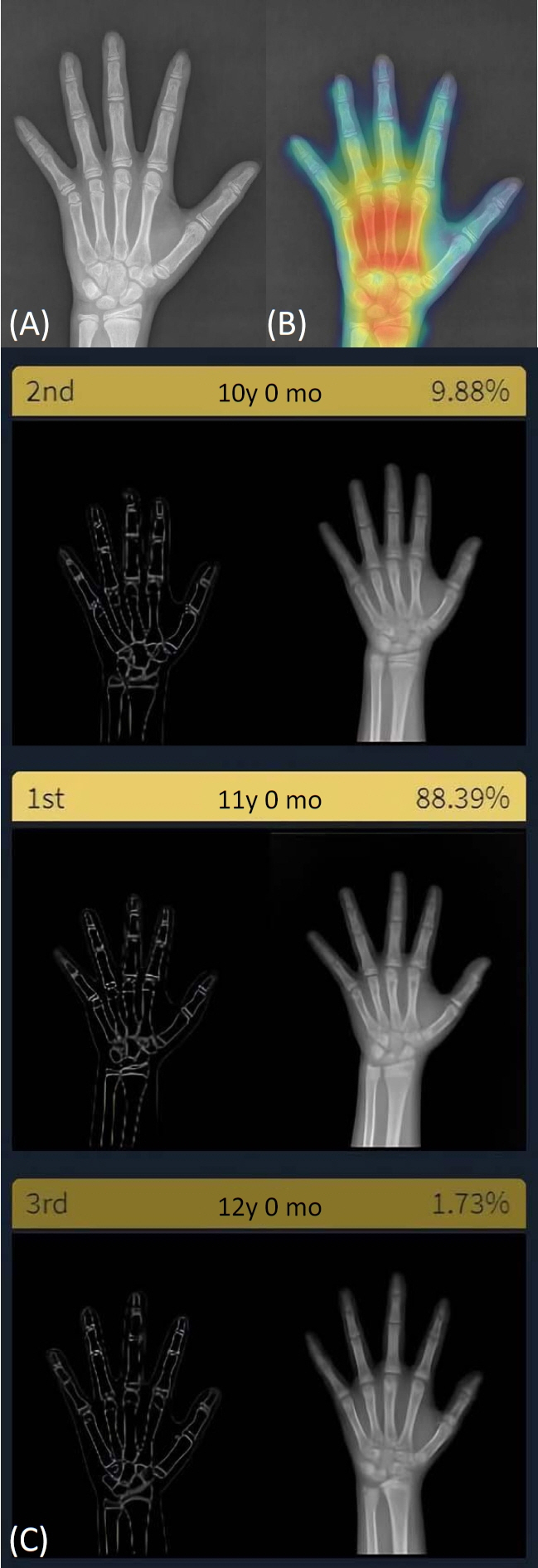
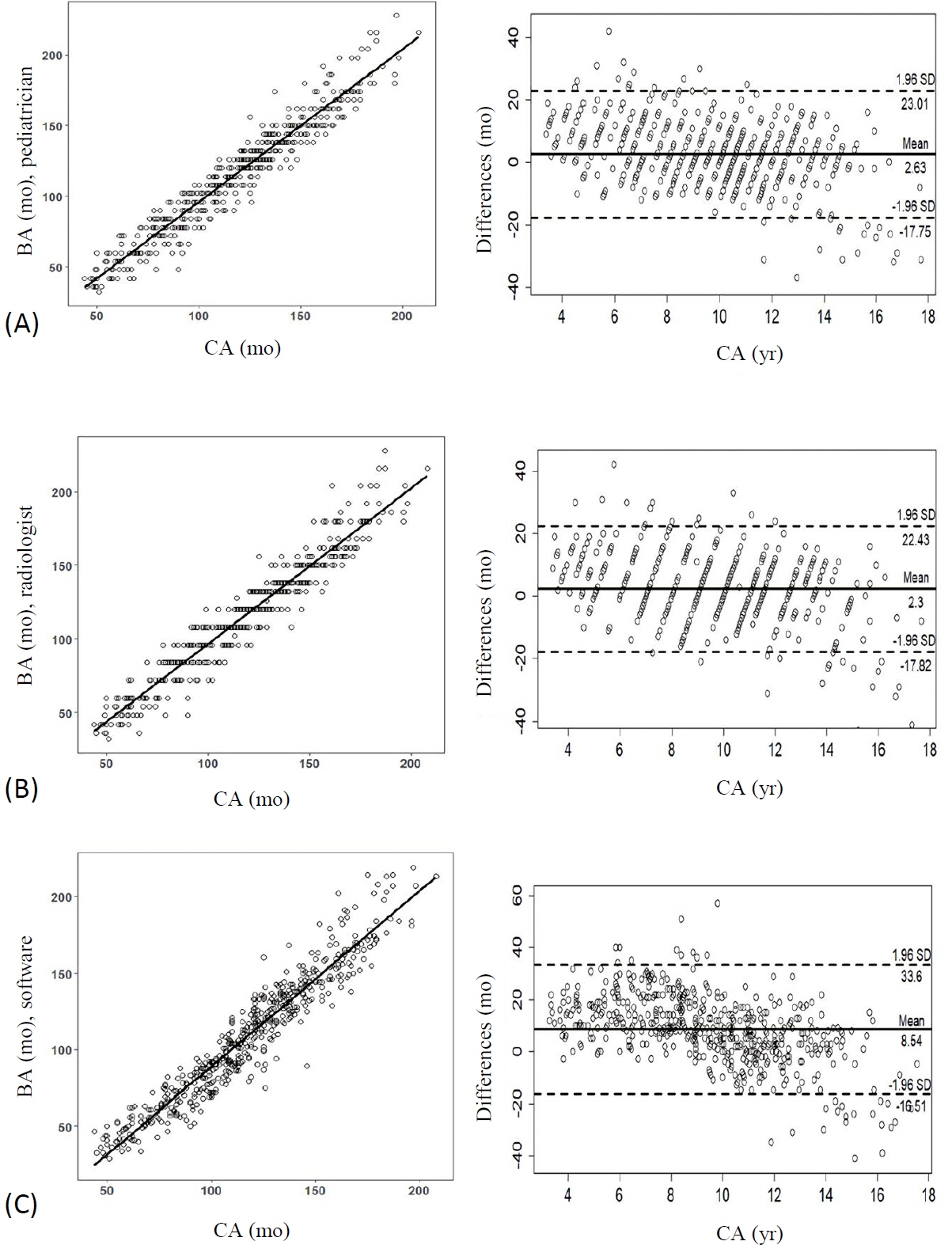
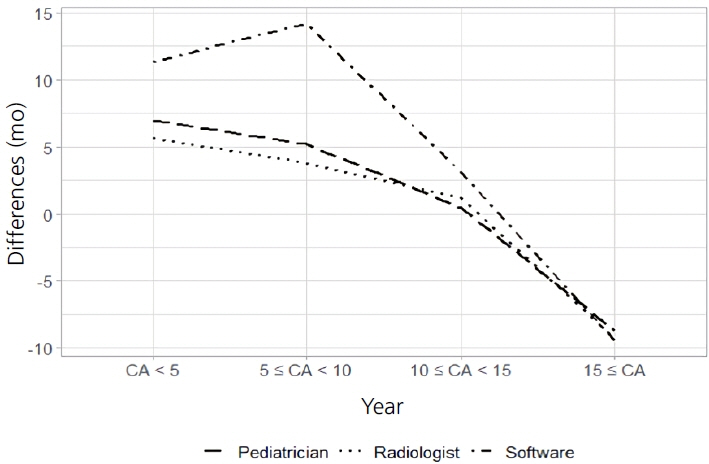
A 2-sample t-test (P<0.001) and Fisher exact test (P=0.011) showed a significant difference between the normal CA and the BA estimated by the DL software. There was good correlation between the 2 variables (r=0.96, P<0.001); however, the root mean square error was 15.4 months. With a 12-month cutoff, the concordance rate was 58.8%. The Bland-Altman plot showed that the DL software tended to underestimate the BA compared with the CA, especially in children under the age of 8.3 years.
Conclusion
The DL-based BA software showed a low concordance rate and a tendency to underestimate the BA in healthy Korean children.
Figure
Reference
-
References
1. Satoh M. Bone age: assessment methods and clinical applications. Clin Pediatr Endocrinol. 2015; 24:143–52.
Article2. Chartrand G, Cheng PM, Vorontsov E, Drozdzal M, Turcotte S, Pal CJ, et al. Deep learning: a primer for radiologists. Radiographics. 2017; 37:2113–31.
Article3. Booz C, Yel I, Wichmann JL, Boettger S, Al Kamali A, Albrecht MH, et al. Artificial intelligence in bone age assessment: accuracy and efficiency of a novel fully automated algorithm compared to the Greulich-Pyle method. Eur Radiol Exp. 2020; 4:6.
Article4. Greulich WW, Pyle SI. Radiographic atlas of skeletal development of the hand and wrist. 2nd ed. Stanford (CA): Stanford University Press;1959.5. Tanner JM, Healy MJR, Goldstein H, Cameron N. Assessment of skeletal maturity and prediction of adult height (TW3 method). 3rd ed. London: W.B. Saunders;2001.6. Hao PY, Chokuwa S, Xie XH, Wu FL, Wu J, Bai C. Skeletal bone age assessments for young children based on regression convolutional neural networks. Math Biosci Eng. 2019; 16:6454–66.
Article7. Chai T, Draxler RR. Root mean square error (RMSE) or mean absolute error (MAE)? - Arguments against avoiding RMSE in the literature. Geosci Model Dev. 2014; 7:1247–50.
Article8. Ramachandran N, Hong SC, Sime MJ, Wilson GA. Diabetic retinopathy screening using deep neural network. Clin Exp Ophthalmol. 2018; 46:412–6.
Article9. Masood A, Sheng B, Li P, Hou X, Wei X, Qin J, et al. Computer-assisted decision support system in pulmonary cancer detection and stage classification on CT images. J Biomed Inform. 2018; 79:117–28.
Article10. Yoo YJ, Ha EJ, Cho YJ, Kim HL, Han M, Kang SY. Computer-aided diagnosis of thyroid nodules via ultrasonography: initial clinical experience. Korean J Radiol. 2018; 19:665–72.
Article11. Polidori N, Castorani V, Mohn A, Chiarelli F. Deciphering short stature in children. Ann Pediatr Endocrinol Metab. 2020; 25:69–79.
Article12. Yue S, Whalen P, Jee YH. Genetic regulation of linear growth. Ann Pediatr Endocrinol Metab. 2019; 24:2–14.
Article13. Cameriere R, De Luca S, Biagi R, Cingolani M, Farronato G, Ferrante L. Accuracy of three age estimation methods in children by measurements of developing teeth and carpals and epiphyses of the ulna and radius. J Forensic Sci. 2012; 57:1263–70.
Article14. Kim JR, Lee YS, Yu J. Assessment of bone age in prepubertal healthy Korean children: comparison among the Korean standard bone age chart, Greulich-Pyle method, and Tanner-Whitehouse method. Korean J Radiol. 2015; 16:201–5.
Article15. Larson DB, Chen MC, Lungren MP, Halabi SS, Stence NV, Langlotz CP. Performance of a deep-learning neural network model in assessing skeletal maturity on pediatric hand radiographs. Radiology. 2018; 287:313–22.
Article16. Lee H, Tajmir S, Lee J, Zissen M, Yeshiwas BA, Alkasab TK, et al. Fully automated deep learning system for bone age assessment. J Digit Imaging. 2017; 30:427–41.
Article17. Kim JR, Shim WH, Yoon HM, Hong SH, Lee JS, Cho YA, et al. Computerized bone age estimation using deep learning based program: evaluation of the accuracy and efficiency. AJR Am J Roentgenol. 2017; 209:1374–80.
Article18. Anthimopoulos M, Christodoulidis S, Ebner L, Christe A, Mougiakakou S. Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans Med Imaging. 2016; 35:1207–16.
Article19. Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017; 284:574–82.
Article20. Boeyer ME, Leary EV, Sherwood RJ, Duren DL. Evidence of the non-linear nature of skeletal maturation. Arch Dis Child. 2020; 105:631–8.
Article21. Eitel KB, Eugster EA. Differences in bone age readings between pediatric endocrinologists and radiologists. Endocr Pract. 2020; 26:328–31.
Article22. Zhang J, Lin F, Ding X. Maturation disparity between hand-wrist bones in a chinese sample of normal children: an analysis based on automatic BoneXpert and manual Greulich and Pyle atlas assessment. Korean J Radiol. 2016; 17:435–42.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Deep-Learning-Based Molecular Imaging Biomarkers: Toward Data-Driven Theranostics
- Development of an Optimized Deep Learning Model for Medical Imaging
- Applying Deep Learning in Medical Images: The Case of Bone Age Estimation
- Deep Learning in Dental Radiographic Imaging
- Bone Age Estimation and Prediction of Final Adult Height Using Deep Learning